A Regression-Based Analysis of Ribosome-Profiling Data Reveals a Conserved Complexity to Mammalian Translation
- PMID: 26638175
- PMCID: PMC4720255
- DOI: 10.1016/j.molcel.2015.11.013
A Regression-Based Analysis of Ribosome-Profiling Data Reveals a Conserved Complexity to Mammalian Translation
Abstract
A fundamental goal of genomics is to identify the complete set of expressed proteins. Automated annotation strategies rely on assumptions about protein-coding sequences (CDSs), e.g., they are conserved, do not overlap, and exceed a minimum length. However, an increasing number of newly discovered proteins violate these rules. Here we present an experimental and analytical framework, based on ribosome profiling and linear regression, for systematic identification and quantification of translation. Application of this approach to lipopolysaccharide-stimulated mouse dendritic cells and HCMV-infected human fibroblasts identifies thousands of novel CDSs, including micropeptides and variants of known proteins, that bear the hallmarks of canonical translation and exhibit translation levels and dynamics comparable to that of annotated CDSs. Remarkably, many translation events are identified in both mouse and human cells even when the peptide sequence is not conserved. Our work thus reveals an unexpected complexity to mammalian translation suited to provide both conserved regulatory or protein-based functions.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures

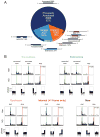
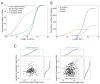
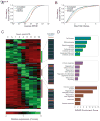

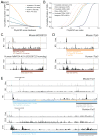
References
-
- Acland P, Dixon M, Peters G, Dickson C. Subcellular fate of the lnt-2 oncoprotein is determined by choice of initiation codon. Nature. 1990;343:662–665. - PubMed
-
- Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet. 2014;15:193–204. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

