mRNA Levels of Imprinted Genes in Bovine In Vivo Oocytes, Embryos and Cross Species Comparisons with Humans, Mice and Pigs
- PMID: 26638780
- PMCID: PMC4671149
- DOI: 10.1038/srep17898
mRNA Levels of Imprinted Genes in Bovine In Vivo Oocytes, Embryos and Cross Species Comparisons with Humans, Mice and Pigs
Abstract
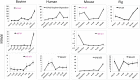
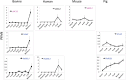
Twenty-six imprinted genes were quantified in bovine in vivo produced oocytes and embryos using RNA-seq. Eighteen were detectable and their transcriptional patterns were: largely decreased (MEST and PLAGL1); first decreased and then increased (CDKN1C and IGF2R); peaked at a specific stage (PHLDA2, SGCE, PEG10, PEG3, GNAS, MEG3, DGAT1, ASCL2, NNAT, and NAP1L5); or constantly low (DIRAS3, IGF2, H19 and RTL1). These patterns reflect mRNAs that are primarily degraded, important at a specific stage, or only required at low quantities. The mRNAs for several genes were surprisingly abundant. For instance, transcripts for the maternally imprinted MEST and PLAGL1, were high in oocytes and could only be expressed from the maternal allele suggesting that their genomic imprints were not yet established/recognized. Although the mRNAs detected here were likely biallelically transcribed before the establishment of imprinted expression, the levels of mRNA during these critical stages of development have important functional consequences. Lastly, we compared these genes to their counterparts in mice, humans and pigs. Apart from previously known differences in the imprinting status, the mRNA levels were different among these four species. The data presented here provide a solid reference for expression profiles of imprinted genes in embryos produced using assisted reproductive biotechnologies.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

