Comparison of Accuracy of Whole-Exome Sequencing with Formalin-Fixed Paraffin-Embedded and Fresh Frozen Tissue Samples
- PMID: 26641479
- PMCID: PMC4671711
- DOI: 10.1371/journal.pone.0144162
Comparison of Accuracy of Whole-Exome Sequencing with Formalin-Fixed Paraffin-Embedded and Fresh Frozen Tissue Samples
Abstract
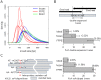
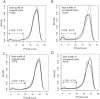
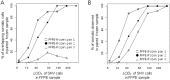
Formalin fixing with paraffin embedding (FFPE) has been a standard sample preparation method for decades, and archival FFPE samples are still very useful resources. Nonetheless, the use of FFPE samples in cancer genome analysis using next-generation sequencing, which is a powerful technique for the identification of genomic alterations at the nucleotide level, has been challenging due to poor DNA quality and artificial sequence alterations. In this study, we performed whole-exome sequencing of matched frozen samples and FFPE samples of tissues from 4 cancer patients and compared the next-generation sequencing data obtained from these samples. The major differences between data obtained from the 2 types of sample were the shorter insert size and artificial base alterations in the FFPE samples. A high proportion of short inserts in the FFPE samples resulted in overlapping paired reads, which could lead to overestimation of certain variants; >20% of the inserts in the FFPE samples were double sequenced. A large number of soft clipped reads was found in the sequencing data of the FFPE samples, and about 30% of total bases were soft clipped. The artificial base alterations, C>T and G>A, were observed in FFPE samples only, and the alteration rate ranged from 200 to 1,200 per 1M bases when sequencing errors were removed. Although high-confidence mutation calls in the FFPE samples were compatible to that in the frozen samples, caution should be exercised in terms of the artifacts, especially for low-confidence calls. Despite the clearly observed artifacts, archival FFPE samples can be a good resource for discovery or validation of biomarkers in cancer research based on whole-exome sequencing.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

