A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae
- PMID: 26641531
- PMCID: PMC4913862
- DOI: 10.1038/nbt.3439
A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae
Abstract
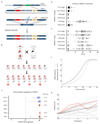
Gene drive systems that enable super-Mendelian inheritance of a transgene have the potential to modify insect populations over a timeframe of a few years. We describe CRISPR-Cas9 endonuclease constructs that function as gene drive systems in Anopheles gambiae, the main vector for malaria. We identified three genes (AGAP005958, AGAP011377 and AGAP007280) that confer a recessive female-sterility phenotype upon disruption, and inserted into each locus CRISPR-Cas9 gene drive constructs designed to target and edit each gene. For each targeted locus we observed a strong gene drive at the molecular level, with transmission rates to progeny of 91.4 to 99.6%. Population modeling and cage experiments indicate that a CRISPR-Cas9 construct targeting one of these loci, AGAP007280, meets the minimum requirement for a gene drive targeting female reproduction in an insect population. These findings could expedite the development of gene drives to suppress mosquito populations to levels that do not support malaria transmission.
Conflict of interest statement
Competing Financial Interests Statement
There are no competing financial interests
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

