The quantitative and condition-dependent Escherichia coli proteome
- PMID: 26641532
- PMCID: PMC4888949
- DOI: 10.1038/nbt.3418
The quantitative and condition-dependent Escherichia coli proteome
Abstract
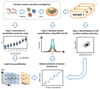
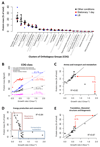
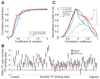
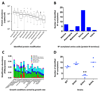
Measuring precise concentrations of proteins can provide insights into biological processes. Here we use efficient protein extraction and sample fractionation, as well as state-of-the-art quantitative mass spectrometry techniques to generate a comprehensive, condition-dependent protein-abundance map for Escherichia coli. We measure cellular protein concentrations for 55% of predicted E. coli genes (>2,300 proteins) under 22 different experimental conditions and identify methylation and N-terminal protein acetylations previously not known to be prevalent in bacteria. We uncover system-wide proteome allocation, expression regulation and post-translational adaptations. These data provide a valuable resource for the systems biology and broader E. coli research communities.
Conflict of interest statement
The authors declare that they have no competing interests as defined by Nature Publishing Group, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
Figures

References
-
- Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

