DYRK1A Controls HIV-1 Replication at a Transcriptional Level in an NFAT Dependent Manner
- PMID: 26641855
- PMCID: PMC4979971
- DOI: 10.1371/journal.pone.0144229
DYRK1A Controls HIV-1 Replication at a Transcriptional Level in an NFAT Dependent Manner
Abstract
Background: Transcription of the HIV-1 provirus is regulated by both viral and host proteins and is very important in the context of viral latency. In latently infected cells, viral gene expression is inhibited as a result of the sequestration of host transcription factors and epigenetic modifications.
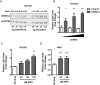
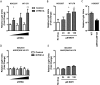
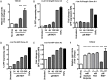
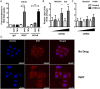
Results: In our present study we analyzed the effect of host factor dual specificity tyrosine-phosphorylation-regulated kinase 1A (DYRK1A) on HIV-1 replication. We show that DYRK1A controls HIV-1 replication by regulating provirus transcription. Downregulation or inhibition of DYRK1A increased LTR-driven transcription and viral replication in cell lines and primary PBMC. Furthermore, inhibition of DYRK1A resulted in reactivation of latent HIV-1 provirus to a similar extent as two commonly used broad-spectrum HDAC inhibitors. We observed that DYRK1A regulates HIV-1 transcription via the Nuclear Factor of Activated T-cells (NFAT) by promoting its translocation from the nucleus to the cytoplasm. Therefore, inhibition of DYRK1A results in increased nuclear levels of NFAT and increased NFAT binding to the viral LTR and thus increasing viral transcription.
Conclusions: Our data indicate that host factor DYRK1A plays a role in the regulation of viral transcription and latency. Therefore, DYRK1A might be an attractive candidate for therapeutic strategies targeting the viral reservoir.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

