Molecular mechanism of respiratory syncytial virus fusion inhibitors
- PMID: 26641933
- PMCID: PMC4731865
- DOI: 10.1038/nchembio.1982
Molecular mechanism of respiratory syncytial virus fusion inhibitors
Abstract
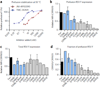
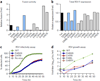
Respiratory syncytial virus (RSV) is a leading cause of pneumonia and bronchiolitis in young children and the elderly. Therapeutic small molecules have been developed that bind the RSV F glycoprotein and inhibit membrane fusion, yet their binding sites and molecular mechanisms of action remain largely unknown. Here we show that these inhibitors bind to a three-fold-symmetric pocket within the central cavity of the metastable prefusion conformation of RSV F. Inhibitor binding stabilizes this conformation by tethering two regions that must undergo a structural rearrangement to facilitate membrane fusion. Inhibitor-escape mutations occur in residues that directly contact the inhibitors or are involved in the conformational rearrangements required to accommodate inhibitor binding. Resistant viruses do not propagate as well as wild-type RSV in vitro, indicating a fitness cost for inhibitor escape. Collectively, these findings provide new insight into class I viral fusion proteins and should facilitate development of optimal RSV fusion inhibitors.
Figures


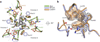
References
-
- Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. Respiratory syncytial virus infection in elderly and high-risk adults. N. Engl. J. Med. 2005;352:1749–1759. - PubMed
-
- The IMpact-RSV Study Group. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics. 1998;102:531–537. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

