Abundant contribution of short tandem repeats to gene expression variation in humans
- PMID: 26642241
- PMCID: PMC4909355
- DOI: 10.1038/ng.3461
Abundant contribution of short tandem repeats to gene expression variation in humans
Abstract
The contribution of repetitive elements to quantitative human traits is largely unknown. Here we report a genome-wide survey of the contribution of short tandem repeats (STRs), which constitute one of the most polymorphic and abundant repeat classes, to gene expression in humans. Our survey identified 2,060 significant expression STRs (eSTRs). These eSTRs were replicable in orthogonal populations and expression assays. We used variance partitioning to disentangle the contribution of eSTRs from that of linked SNPs and indels and found that eSTRs contribute 10-15% of the cis heritability mediated by all common variants. Further functional genomic analyses showed that eSTRs are enriched in conserved regions, colocalize with regulatory elements and may modulate certain histone modifications. By analyzing known genome-wide association study (GWAS) signals and searching for new associations in 1,685 whole genomes from deeply phenotyped individuals, we found that eSTRs are enriched in various clinically relevant conditions. These results highlight the contribution of STRs to the genetic architecture of quantitative human traits.
Figures

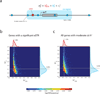
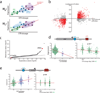

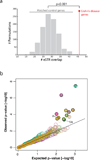
Comment in
-
Complex traits: Repeat, repeat, repeat--gene expression variability explained.Nat Rev Genet. 2016 Feb;17(2):68-9. doi: 10.1038/nrg.2015.27. Epub 2015 Dec 30. Nat Rev Genet. 2016. PMID: 26715326 No abstract available.
References
-
- Moffatt MF, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 1U01HG007037/HG/NHGRI NIH HHS/United States
- U01 HG007037/HG/NHGRI NIH HHS/United States
- R01 HG006399/HG/NHGRI NIH HHS/United States
- R01MH084703/MH/NIMH NIH HHS/United States
- DA033660/DA/NIDA NIH HHS/United States
- R01 MH097018/MH/NIMH NIH HHS/United States
- R01 MH084703/MH/NIMH NIH HHS/United States
- S10 OD018522/OD/NIH HHS/United States
- R01 HG006696/HG/NHGRI NIH HHS/United States
- MH097018/MH/NIMH NIH HHS/United States
- R01 DA033660/DA/NIDA NIH HHS/United States
- R01HG006399/HG/NHGRI NIH HHS/United States
- HG006696/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

