Comparative genomics of Pseudomonas fluorescens subclade III strains from human lungs
- PMID: 26644001
- PMCID: PMC4672498
- DOI: 10.1186/s12864-015-2261-2
Comparative genomics of Pseudomonas fluorescens subclade III strains from human lungs
Abstract
Background: While the taxonomy and genomics of environmental strains from the P. fluorescens species-complex has been reported, little is known about P. fluorescens strains from clinical samples. In this report, we provide the first genomic analysis of P. fluorescens strains in which human vs. environmental isolates are compared.
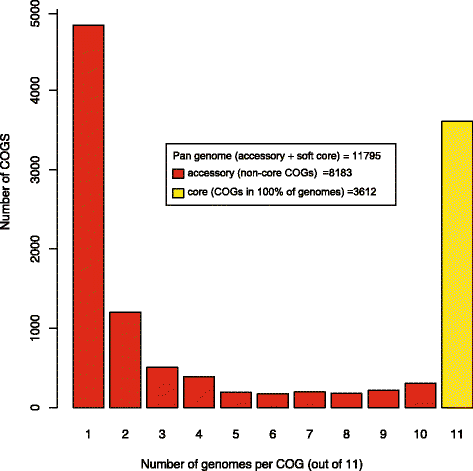
Results: Seven P. fluorescens strains were isolated from respiratory samples from cystic fibrosis (CF) patients. The clinical strains could grow at a higher temperature (>34 °C) than has been reported for environmental strains. Draft genomes were generated for all of the clinical strains, and multi-locus sequence analysis placed them within subclade III of the P. fluorescens species-complex. All strains encoded type- II, -III, -IV, and -VI secretion systems, as well as the widespread colonization island (WCI). This is the first description of a WCI in P. fluorescens strains. All strains also encoded a complete I2/PfiT locus and showed evidence of horizontal gene transfer. The clinical strains were found to differ from the environmental strains in the number of genes involved in metal resistance, which may be a possible adaptation to chronic antibiotic exposure in the CF lung.
Conclusions: This is the largest comparative genomics analysis of P. fluorescens subclade III strains to date and includes the first clinical isolates. At a global level, the clinical P. fluorescens subclade III strains were largely indistinguishable from environmental P. fluorescens subclade III strains, supporting the idea that identifying strains as 'environmental' vs 'clinical' is not a phenotypic trait. Rather, strains within P. fluorescens subclade III will colonize and persist in any niche that provides the requirements necessary for growth.
Figures






Similar articles
-
Type III secretion system and virulence markers highlight similarities and differences between human- and plant-associated pseudomonads related to Pseudomonas fluorescens and P. putida.Appl Environ Microbiol. 2015 Apr;81(7):2579-90. doi: 10.1128/AEM.04160-14. Epub 2015 Jan 30. Appl Environ Microbiol. 2015. PMID: 25636837 Free PMC article.
-
Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions.PLoS Genet. 2012 Jul;8(7):e1002784. doi: 10.1371/journal.pgen.1002784. Epub 2012 Jul 5. PLoS Genet. 2012. PMID: 22792073 Free PMC article.
-
Genomic analysis of the biocontrol strain Pseudomonas fluorescens Pf29Arp with evidence of T3SS and T6SS gene expression on plant roots.Environ Microbiol Rep. 2013 Jun;5(3):393-403. doi: 10.1111/1758-2229.12048. Epub 2013 Apr 10. Environ Microbiol Rep. 2013. PMID: 23754720
-
Genomic and Genetic Diversity within the Pseudomonas fluorescens Complex.PLoS One. 2016 Feb 25;11(2):e0150183. doi: 10.1371/journal.pone.0150183. eCollection 2016. PLoS One. 2016. PMID: 26915094 Free PMC article.
-
Microbiology, genomics, and clinical significance of the Pseudomonas fluorescens species complex, an unappreciated colonizer of humans.Clin Microbiol Rev. 2014 Oct;27(4):927-48. doi: 10.1128/CMR.00044-14. Clin Microbiol Rev. 2014. PMID: 25278578 Free PMC article. Review.
Cited by
-
Potential Biocontrol Activities of Populus Endophytes against Several Plant Pathogens Using Different Inhibitory Mechanisms.Pathogens. 2022 Dec 22;12(1):13. doi: 10.3390/pathogens12010013. Pathogens. 2022. PMID: 36678361 Free PMC article.
-
The oral microbiota - a mechanistic role for systemic diseases.Br Dent J. 2018 Mar 23;224(6):447-455. doi: 10.1038/sj.bdj.2018.217. Br Dent J. 2018. PMID: 29569607 Review.
-
Whole-Genome Sequencing-Based Re-Identification of Pseudomonas putida/fluorescens Clinical Isolates Identified by Biochemical Bacterial Identification Systems.Microbiol Spectr. 2022 Apr 27;10(2):e0249121. doi: 10.1128/spectrum.02491-21. Epub 2022 Apr 7. Microbiol Spectr. 2022. PMID: 35389240 Free PMC article.
-
Pneumonia caused by Pseudomonas fluorescens: a case report.BMC Pulm Med. 2021 Jul 5;21(1):212. doi: 10.1186/s12890-021-01573-9. BMC Pulm Med. 2021. PMID: 34225696 Free PMC article.
-
Determining the Different Mechanisms Used by Pseudomonas Species to Cope With Minimal Inhibitory Concentrations of Zinc via Comparative Transcriptomic Analyses.Front Microbiol. 2020 Dec 3;11:573857. doi: 10.3389/fmicb.2020.573857. eCollection 2020. Front Microbiol. 2020. PMID: 33343517 Free PMC article.
References
-
- Loper JE, Hassan KA, Mavrodi DV, Davis EW, 2nd, Lim CK, Shaffer BT, et al. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 2012;8(7):e1002784. doi: 10.1371/journal.pgen.1002784. - DOI - PMC - PubMed
-
- Mulet M, Lalucat J, Garcia-Valdes E. DNA sequence-based analysis of the Pseudomonas species. Environ Microbiol. 2010;12(6):1513–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

