Deregulated KLF4 Expression in Myeloid Leukemias Alters Cell Proliferation and Differentiation through MicroRNA and Gene Targets
- PMID: 26644403
- PMCID: PMC4751692
- DOI: 10.1128/MCB.00712-15
Deregulated KLF4 Expression in Myeloid Leukemias Alters Cell Proliferation and Differentiation through MicroRNA and Gene Targets
Abstract
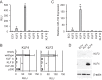
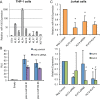
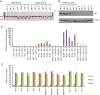
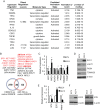
Acute myeloid leukemia (AML) is characterized by increased proliferation and blocked differentiation of hematopoietic progenitors mediated, in part, by altered myeloid transcription factor expression. Decreased Krüppel-like factor 4 (KLF4) expression has been observed in AML, but how decreased KLF4 contributes to AML pathogenesis is largely unknown. We demonstrate decreased KLF4 expression in AML patient samples with various cytogenetic aberrations, confirm that KLF4 overexpression promotes myeloid differentiation and inhibits cell proliferation in AML cell lines, and identify new targets of KLF4. We have demonstrated that microRNA 150 (miR-150) expression is decreased in AML and that reintroducing miR-150 expression induces myeloid differentiation and inhibits proliferation of AML cells. We show that KLF family DNA binding sites are necessary for miR-150 promoter activity and that KLF2 or KLF4 overexpression induces miR-150 expression. miR-150 silencing, alone or in combination with silencing of CDKN1A, a well-described KLF4 target, did not fully reverse KLF4-mediated effects. Gene expression profiling and validation identified putative KLF4-regulated genes, including decreased MYC and downstream MYC-regulated gene expression in KLF4-overexpressing cells. Our findings indicate that decreased KLF4 expression mediates antileukemic effects through regulation of gene and microRNA networks, containing miR-150, CDKN1A, and MYC, and provide mechanistic support for therapeutic strategies increasing KLF4 expression.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures






Similar articles
-
HDAC1 and Klf4 interplay critically regulates human myeloid leukemia cell proliferation.Cell Death Dis. 2014 Oct 23;5(10):e1491. doi: 10.1038/cddis.2014.433. Cell Death Dis. 2014. PMID: 25341045 Free PMC article.
-
Overexpression of KLF4 promotes cell senescence through microRNA-203-survivin-p21 pathway.Oncotarget. 2016 Sep 13;7(37):60290-60302. doi: 10.18632/oncotarget.11200. Oncotarget. 2016. PMID: 27531889 Free PMC article.
-
A novel miR-375-HOXB3-CDCA3/DNMT3B regulatory circuitry contributes to leukemogenesis in acute myeloid leukemia.BMC Cancer. 2018 Feb 13;18(1):182. doi: 10.1186/s12885-018-4097-z. BMC Cancer. 2018. PMID: 29439669 Free PMC article.
-
MicroRNA expression in acute myeloid leukemia.Curr Hematol Malig Rep. 2009 Apr;4(2):83-8. doi: 10.1007/s11899-009-0012-7. Curr Hematol Malig Rep. 2009. PMID: 20425419 Review.
-
MicroRNAs expressed in hematopoietic stem/progenitor cells are deregulated in acute myeloid leukemias.Leuk Lymphoma. 2015 May;56(5):1466-74. doi: 10.3109/10428194.2014.955019. Epub 2015 Jan 21. Leuk Lymphoma. 2015. PMID: 25242094 Review.
Cited by
-
Molecular subtyping of acute myeloid leukemia through ferroptosis signatures predicts prognosis and deciphers the immune microenvironment.Front Cell Dev Biol. 2023 Aug 24;11:1207642. doi: 10.3389/fcell.2023.1207642. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37691822 Free PMC article.
-
Krüppel-Like Factor 4 Enhances Sensitivity of Cisplatin to Lung Cancer Cells and Inhibits Regulating Epithelial-to-Mesenchymal Transition.Oncol Res. 2016;24(2):81-7. doi: 10.3727/096504016X14597766487717. Oncol Res. 2016. PMID: 27296948 Free PMC article.
-
miR-190a-5p participates in the regulation of hypoxia-induced pulmonary hypertension by targeting KLF15 and can serve as a biomarker of diagnosis and prognosis in chronic obstructive pulmonary disease complicated with pulmonary hypertension.Int J Chron Obstruct Pulmon Dis. 2018 Nov 20;13:3777-3790. doi: 10.2147/COPD.S182504. eCollection 2018. Int J Chron Obstruct Pulmon Dis. 2018. PMID: 30538440 Free PMC article.
-
Novel tumor-suppressor function of KLF4 in pediatric T-cell acute lymphoblastic leukemia.Exp Hematol. 2017 Sep;53:16-25. doi: 10.1016/j.exphem.2017.04.009. Epub 2017 May 4. Exp Hematol. 2017. PMID: 28479419 Free PMC article. Review.
-
Subtype-specific 3D genome alteration in acute myeloid leukaemia.Nature. 2022 Nov;611(7935):387-398. doi: 10.1038/s41586-022-05365-x. Epub 2022 Oct 26. Nature. 2022. PMID: 36289338 Free PMC article.
References
-
- Rosenbauer F, Tenen DG. 2007. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol 7:105–117. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
