Disulfide-Bond-Forming Pathways in Gram-Positive Bacteria
- PMID: 26644434
- PMCID: PMC4810614
- DOI: 10.1128/JB.00769-15
Disulfide-Bond-Forming Pathways in Gram-Positive Bacteria
Abstract
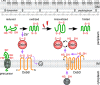
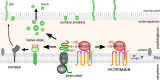
Disulfide bonds are important for the stability and function of many secreted proteins. In Gram-negative bacteria, these linkages are catalyzed by thiol-disulfide oxidoreductases (Dsb) in the periplasm. Protein oxidation has been well studied in these organisms, but it has not fully been explored in Gram-positive bacteria, which lack traditional periplasmic compartments. Recent bioinformatics analyses have suggested that the high-GC-content bacteria (i.e., actinobacteria) rely on disulfide-bond-forming pathways. In support of this, Dsb-like proteins have been identified in Mycobacterium tuberculosis, but their functions are not known. Actinomyces oris and Corynebacterium diphtheriae have recently emerged as models to study disulfide bond formation in actinobacteria. In both organisms, disulfide bonds are catalyzed by the membrane-bound oxidoreductase MdbA. Remarkably, unlike known Dsb proteins, MdbA is important for pathogenesis and growth, which makes it a potential target for new antibacterial drugs. This review will discuss disulfide-bond-forming pathways in bacteria, with a special focus on Gram-positive bacteria.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
References
-
- Goldberger RF, Epstein CJ, Anfinsen CB. 1963. Acceleration of reactivation of reduced bovine pancreatic ribonuclease by a microsomal system from rat liver. J Biol Chem 238:628–635. - PubMed
-
- Givol D, Goldberger RF, Anfinsen CB. 1964. Oxidation and disulfide interchange in the reactivation of reduced ribonuclease. J Biol Chem 239:PC3114–PC3116. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

