Binding of the Covalent Flavin Assembly Factor to the Flavoprotein Subunit of Complex II
- PMID: 26644464
- PMCID: PMC4742753
- DOI: 10.1074/jbc.M115.690396
Binding of the Covalent Flavin Assembly Factor to the Flavoprotein Subunit of Complex II
Abstract
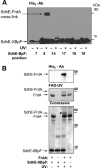
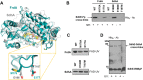
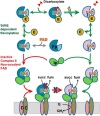
Escherichia coli harbors two highly conserved homologs of the essential mitochondrial respiratory complex II (succinate:ubiquinone oxidoreductase). Aerobically the bacterium synthesizes succinate:quinone reductase as part of its respiratory chain, whereas under microaerophilic conditions, the quinol:fumarate reductase can be utilized. All complex II enzymes harbor a covalently bound FAD co-factor that is essential for their ability to oxidize succinate. In eukaryotes and many bacteria, assembly of the covalent flavin linkage is facilitated by a small protein assembly factor, termed SdhE in E. coli. How SdhE assists with formation of the covalent flavin bond and how it binds the flavoprotein subunit of complex II remain unknown. Using photo-cross-linking, we report the interaction site between the flavoprotein of complex II and the SdhE assembly factor. These data indicate that SdhE binds to the flavoprotein between two independently folded domains and that this binding mode likely influences the interdomain orientation. In so doing, SdhE likely orients amino acid residues near the dicarboxylate and FAD binding site, which facilitates formation of the covalent flavin linkage. These studies identify how the conserved SdhE assembly factor and its homologs participate in complex II maturation.
Keywords: chaperone; complex II; flavin adenine dinucleotide (FAD); fumarate reductase; mitochondrial respiratory chain complex; protein assembly; protein self-assembly; protein-protein interaction; succinate dehydrogenase.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






References
-
- Cecchini G. (2003) Function and structure of complex II of the respiratory chain. Annu. Rev. Biochem. 72, 77–109 - PubMed
-
- Iverson T. M., Luna-Chavez C., Cecchini G., and Rees D. C. (1999) Structure of the Escherichia coli fumarate reductase respiratory complex. Science 284, 1961–1966 - PubMed
-
- Lancaster C. R., Kröger A., Auer M., and Michel H. (1999) Structure of fumarate reductase from Wolinella succinogenes at 2.2 A resolution. Nature 402, 377–385 - PubMed
-
- Yankovskaya V., Horsefield R., Törnroth S., Luna-Chavez C., Miyoshi H., Léger C., Byrne B., Cecchini G., and Iwata S. (2003) Architecture of succinate dehydrogenase and reactive oxygen species generation. Science 299, 700–704 - PubMed
-
- Sun F., Huo X., Zhai Y., Wang A., Xu J., Su D., Bartlam M., and Rao Z. (2005) Crystal structure of mitochondrial respiratory membrane protein complex II. Cell 121, 1043–1057 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

