A Molecular Host Response Assay to Discriminate Between Sepsis and Infection-Negative Systemic Inflammation in Critically Ill Patients: Discovery and Validation in Independent Cohorts
- PMID: 26645559
- PMCID: PMC4672921
- DOI: 10.1371/journal.pmed.1001916
A Molecular Host Response Assay to Discriminate Between Sepsis and Infection-Negative Systemic Inflammation in Critically Ill Patients: Discovery and Validation in Independent Cohorts
Abstract
Background: Systemic inflammation is a whole body reaction having an infection-positive (i.e., sepsis) or infection-negative origin. It is important to distinguish between these two etiologies early and accurately because this has significant therapeutic implications for critically ill patients. We hypothesized that a molecular classifier based on peripheral blood RNAs could be discovered that would (1) determine which patients with systemic inflammation had sepsis, (2) be robust across independent patient cohorts, (3) be insensitive to disease severity, and (4) provide diagnostic utility. The goal of this study was to identify and validate such a molecular classifier.
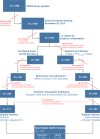
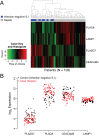
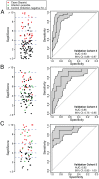
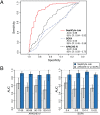
Methods and findings: We conducted an observational, non-interventional study of adult patients recruited from tertiary intensive care units (ICUs). Biomarker discovery utilized an Australian cohort (n = 105) consisting of 74 cases (sepsis patients) and 31 controls (post-surgical patients with infection-negative systemic inflammation) recruited at five tertiary care settings in Brisbane, Australia, from June 3, 2008, to December 22, 2011. A four-gene classifier combining CEACAM4, LAMP1, PLA2G7, and PLAC8 RNA biomarkers was identified. This classifier, designated SeptiCyte Lab, was validated using reverse transcription quantitative PCR and receiver operating characteristic (ROC) curve analysis in five cohorts (n = 345) from the Netherlands. Patients for validation were selected from the Molecular Diagnosis and Risk Stratification of Sepsis study (ClinicalTrials.gov, NCT01905033), which recruited ICU patients from the Academic Medical Center in Amsterdam and the University Medical Center Utrecht. Patients recruited from November 30, 2012, to August 5, 2013, were eligible for inclusion in the present study. Validation cohort 1 (n = 59) consisted entirely of unambiguous cases and controls; SeptiCyte Lab gave an area under curve (AUC) of 0.95 (95% CI 0.91-1.00) in this cohort. ROC curve analysis of an independent, more heterogeneous group of patients (validation cohorts 2-5; 249 patients after excluding 37 patients with an infection likelihood of "possible") gave an AUC of 0.89 (95% CI 0.85-0.93). Disease severity, as measured by Sequential Organ Failure Assessment (SOFA) score or Acute Physiology and Chronic Health Evaluation (APACHE) IV score, was not a significant confounding variable. The diagnostic utility of SeptiCyte Lab was evaluated by comparison to various clinical and laboratory parameters available to a clinician within 24 h of ICU admission. SeptiCyte Lab was significantly better at differentiating cases from controls than all tested parameters, both singly and in various logistic combinations, and more than halved the diagnostic error rate compared to procalcitonin in all tested cohorts and cohort combinations. Limitations of this study relate to (1) cohort compositions that do not perfectly reflect the composition of the intended use population, (2) potential biases that could be introduced as a result of the current lack of a gold standard for diagnosing sepsis, and (3) lack of a complete, unbiased comparison to C-reactive protein.
Conclusions: SeptiCyte Lab is a rapid molecular assay that may be clinically useful in managing ICU patients with systemic inflammation. Further study in population-based cohorts is needed to validate this assay for clinical use.
Conflict of interest statement
The authors have read the journal’s policy, and declare the following: AJS JL PKK LVV BS MB OC MJS TVDP have no competing interests; TAS LM AR JTK TDY RAB RBB DJV MRT are employees and/or shareholders of Immunexpress, which holds the intellectual property in SeptiCyte Lab; JJP has no competing interests except for this: "In 2010 Immunexpress Inc. paid to the Adult Intensive Care Unit, Mater Health Services, Brisbane, Australia a sum of AUS $38,509 (thirty-eight thousand, five hundred and nine Australian dollars) as half-time temporary salary support for an ICU research coordinator to facilitate patient identification, recruitment and sample collection for 6 months from 20 May 2010.”
Figures

References
-
- Calandra T, Cohen J. The International Sepsis Forum consensus conference on definitions of infection in the intensive care unit. Crit Care Med. 2005;33:1538–1548. - PubMed
-
- Dellinger RP, Levy MM, Carlet JM, Bion J, Parker MM, Jaeschke R, et al. Surviving Sepsis Campaign: international guidelines for management of severe sepsis and septic shock. Crit Care Med. 2008;36:296–327. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

