Mechanical sensitivity of Piezo1 ion channels can be tuned by cellular membrane tension
- PMID: 26646186
- PMCID: PMC4718726
- DOI: 10.7554/eLife.12088
Mechanical sensitivity of Piezo1 ion channels can be tuned by cellular membrane tension
Abstract
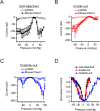
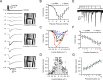
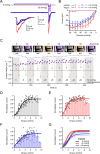
Piezo1 ion channels mediate the conversion of mechanical forces into electrical signals and are critical for responsiveness to touch in metazoans. The apparent mechanical sensitivity of Piezo1 varies substantially across cellular environments, stimulating methods and protocols, raising the fundamental questions of what precise physical stimulus activates the channel and how its stimulus sensitivity is regulated. Here, we measured Piezo1 currents evoked by membrane stretch in three patch configurations, while simultaneously visualizing and measuring membrane geometry. Building on this approach, we developed protocols to minimize resting membrane curvature and tension prior to probing Piezo1 activity. We find that Piezo1 responds to lateral membrane tension with exquisite sensitivity as compared to other mechanically activated channels and that resting tension can drive channel inactivation, thereby tuning overall mechanical sensitivity of Piezo1. Our results explain how Piezo1 can function efficiently and with adaptable sensitivity as a sensor of mechanical stimulation in diverse cellular contexts.
Keywords: Piezo1; biophysics; inactivation; ion channel; mechanically activated; neuroscience; none; structural biology; tension.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures


References
-
- Albuisson J, Murthy SE, Bandell M, Coste B, Louis-Dit-Picard H, Mathur J, Fénéant-Thibault M, Tertian G, de Jaureguiberry JP, Syfuss PY, Cahalan S, Garçon L, Toutain F, Simon Rohrlich P, Delaunay J, Picard V, Jeunemaitre X, Patapoutian A. Dehydrated hereditary stomatocytosis linked to gain-of-function mutations in mechanically activated PIEZO1 ion channels. Nature Communications. 2013;4:1884. doi: 10.1038/ncomms2899. - DOI - PMC - PubMed
-
- Bae C, Gnanasambandam R, Nicolai C, Sachs F, Gottlieb PA. Xerocytosis is caused by mutations that alter the kinetics of the mechanosensitive channel PIEZO1. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:e12088. doi: 10.1073/pnas.1219777110. - DOI - PMC - PubMed
-
- Blumenthal NR, Hermanson O, Heimrich B, Shastri VP. Stochastic nanoroughness modulates neuron–astrocyte interactions and function via mechanosensing cation channels. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:16124–16129. doi: 10.1073/pnas.1412740111. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

