Hormone stimulation of androgen receptor mediates dynamic changes in DNA methylation patterns at regulatory elements
- PMID: 26646795
- PMCID: PMC4767454
- DOI: 10.18632/oncotarget.6471
Hormone stimulation of androgen receptor mediates dynamic changes in DNA methylation patterns at regulatory elements
Abstract
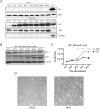
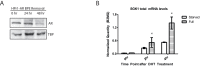
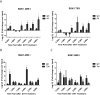
DNA methylation is an epigenetic modification that contributes to stable gene silencing by interfering with the ability of transcriptional regulators to bind to DNA. Recent findings have revealed that hormone stimulation of certain nuclear receptors induces rapid, dynamic changes in DNA methylation patterns alongside transcriptional responses at a subset of target loci, over time. However, the ability of androgen receptor (AR) to dynamically regulate gene transcription is relatively under-studied and its role in the regulation of DNA methylation patterns remains to be elucidated. Here we demonstrate in normal prostate cells that hormone stimulated AR activity results in dynamic changes in the transcription rate and DNA methylation patterns at the AR target genes, TIPARP and SGK1. Time-resolved chromatin immunoprecipitation experiments on the SGK1 locus reveals dynamic recruitment of AR and RNA Polymerase II, as well as the recruitment of proteins involved in the DNA demethylation process, TET1 and TDG. Furthermore, the presence of DNA methylation at dynamic regions inhibits protein binding and transcriptional activity of SGK1. These findings establish AR activity as a contributing factor to the dynamic regulation of DNA methylation patterns at target genes in prostate biology and infer further complexity involved in nuclear receptor mediation of transcriptional regulation.
Keywords: Chromosome Section; DNA methylation; androgen receptor; androgen regulated target genes; gene transcription; nuclear receptor.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

