NMR characterization of a 72 kDa transcription factor using differential isotopic labeling
- PMID: 26647230
- PMCID: PMC4815403
- DOI: 10.1002/pro.2853
NMR characterization of a 72 kDa transcription factor using differential isotopic labeling
Abstract
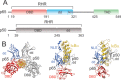
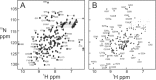
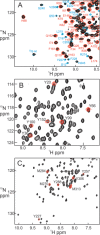
NF-κB is a major transcription factor that mediates a number of cellular signaling pathways. Crystal structure analysis gives an incomplete picture of the behavior of the protein, particularly in the free state; free monomers or dimers of NF-κB have never been crystallized. NMR analysis gives insights into the structure and dynamics of the protein in solution, but a necessary first step is the assignment of resonances. The size of the heterodimer of the Rel homology regions of the NF-κB monomers p65 and p50 (72 kDa) prohibits the straightforward use of triple-resonance spectroscopy to obtain the assignments. However, the dynamic nature of the free heterodimer, in particular the independence of the DNA-binding and dimerization domains of each monomer, allows the assignments made on differentially labeled smaller domains to be mapped successfully onto the spectrum of the larger full-length RHR. Problematic areas such as the p65 nuclear localization sequence, which is disordered in the free protein, can be approached by residue-specific labeling and comparison with previously-published spectra of a short peptide with the same sequence. Overall, this NMR analysis of NF-κB has given valuable insights into the highly dynamic nature of the free state, which is likely to play an important role in the functional cycle of NF-κB in the cell.
Keywords: IκBα; NF-κB; NMR; dynamics; structure.
© 2015 The Protein Society.
Figures

References
-
- Hoffmann A, Natoli G, Ghosh G (2006) Transcriptional regulation via the NF‐kB signaling module. Oncogene 25:6706–6716. - PubMed
-
- Beg AA, Baldwin AS, Jr (1993) The IkB proteins: multifunctional regulators of Rel/NF‐kB transcription factors. Genes Dev 7:2064–2070. - PubMed
-
- Chen FE, Huang DB, Chen YQ, Ghosh G (1998) Crystal structure of p50/p65 heterodimer of transcription factor NF‐kB bound to DNA. Nature 391:410–413. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

