De novo dominant ASXL3 mutations alter H2A deubiquitination and transcription in Bainbridge-Ropers syndrome
- PMID: 26647312
- PMCID: PMC4731023
- DOI: 10.1093/hmg/ddv499
De novo dominant ASXL3 mutations alter H2A deubiquitination and transcription in Bainbridge-Ropers syndrome
Abstract
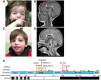
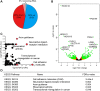
De novo truncating mutations in Additional sex combs-like 3 (ASXL3) have been identified in individuals with Bainbridge-Ropers syndrome (BRS), characterized by failure to thrive, global developmental delay, feeding problems, hypotonia, dysmorphic features, profound speech delays and intellectual disability. We identified three novel de novo heterozygous truncating variants distributed across ASXL3, outside the original cluster of ASXL3 mutations previously described for BRS. Primary skin fibroblasts established from a BRS patient were used to investigate the functional impact of pathogenic variants. ASXL3 mRNA transcripts from the mutated allele are prone to nonsense-mediated decay, and expression of ASXL3 is reduced. We found that ASXL3 interacts with BAP1, a hydrolase that removes mono-ubiquitin from histone H2A lysine 119 (H2AK119Ub1) as a component of the Polycomb repressive deubiquitination (PR-DUB) complex. A significant increase in H2AK119Ub1 was observed in ASXL3 patient fibroblasts, highlighting an important functional role for ASXL3 in PR-DUB mediated deubiquitination. Transcriptomes of ASXL3 patient and control fibroblasts were compared to investigate the impact of chromatin changes on transcriptional regulation. Out of 564 significantly differentially expressed genes (DEGs) in ASXL3 patient fibroblasts, 52% were upregulated and 48% downregulated. DEGs were enriched in molecular processes impacting transcriptional regulation, development and proliferation, consistent with the features of BRS. This is the first single gene disorder linked to defects in deubiquitination of H2AK119Ub1 and suggests an important role for dynamic regulation of H2A mono-ubiquitination in transcriptional regulation and the pathophysiology of BRS.
© The Author 2015. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures


References
-
- Ku C.S., Polychronakos C., Tan E.K., Naidoo N., Pawitan Y., Roukos D.H., Mort M., Cooper D.N. (2013) A new paradigm emerges from the study of de novo mutations in the context of neurodevelopmental disease. Mol. Psychiatry, 18, 141–153. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

