Protease signaling in animal and plant-regulated cell death
- PMID: 26648190
- PMCID: PMC5606204
- DOI: 10.1111/febs.13616
Protease signaling in animal and plant-regulated cell death
Abstract
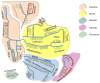
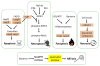
This review aims to highlight the proteases required for regulated cell death mechanisms in animals and plants. The aim is to be incisive, and not inclusive of all the animal proteases that have been implicated in various publications. The review also aims to focus on instances when several publications from disparate groups have demonstrated the involvement of an animal protease, and also when there is substantial biochemical, mechanistic and genetic evidence. In doing so, the literature can be culled to a handful of proteases, covering most of the known regulated cell death mechanisms: apoptosis, regulated necrosis, necroptosis, pyroptosis and NETosis in animals. In plants, the literature is younger and not as extensive as for mammals, although the molecular drivers of vacuolar death, necrosis and the hypersensitive response in plants are becoming clearer. Each of these death mechanisms has at least one proteolytic component that plays a major role in controlling the pathway, and sometimes they combine in networks to regulate cell death/survival decision nodes. Some similarities are found among animal and plant cell death proteases but, overall, the pathways that they govern are kingdom-specific with very little overlap.
Keywords: NETosis; apoptosis; caspase; cathepsin; metacaspase; necroptosis; necrosis; peptidase; proteolysis; pyroptosis.
© 2015 FEBS.
Figures



References
-
- Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, Hengartner M, Knight RA, Kumar S, Lipton SA, Malorni W, Nunez G, Peter ME, Tschopp J, Yuan J, Piacentini M, Zhivotovsky B, Melino G. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. - PMC - PubMed
-
- Galluzzi, Bravo-San Pedro L, Vitale JM, Aaronson I, Abrams SA, Adam JM, Alnemri D, Altucci ES, Andrews L, Annicchiarico-Petruzzelli D, Baehrecke M, Bazan EH, Bertrand NG, Bianchi MJ, Blagosklonny K, Blomgren MV, Borner K, Bredesen C, Brenner DE, Campanella C, Candi M, Cecconi E, Chan F, Chandel FK, Cheng NS, Chipuk EH, Cidlowski JE, Ciechanover JA, Dawson A, Dawson TM, De Laurenzi VL, De Maria V, Debatin R, Di Daniele KM, Dixit N, Dynlacht VM, El-Deiry BD, Fimia WS, Flavell GM, Fulda RA, Garrido S, Gougeon C, Green ML, Gronemeyer DR, Hajnoczky H, Hardwick G, Hengartner JM, Ichijo MO, Joseph H, Jost B, Kaufmann PJ, Kepp T, Klionsky O, Knight DJ, Kumar RA, Lemasters S, Levine JJ, Linkermann B, Lipton A, Lockshin SA, Lopez-Otin RA, Lugli C, Madeo E, Malorni F, Marine W, Martin JC, Martinou SJ, Medema JC, Meier JP, Melino P, Mizushima S, Moll N, Munoz-Pinedo U, Nunez C, Oberst G, Panaretakis A, Penninger T, Peter JM, Piacentini ME, Pinton M, Prehn P, Puthalakath JH, Rabinovich H, Ravichandran GA, Rizzuto KS, Rodrigues R, Rubinsztein CM, Rudel DC, Shi T, Simon Y, Stockwell HU, Szabadkai BR, Tait G, Tang SW, Tavernarakis HL, Tsujimoto N, Vanden Berghe Y, Vandenabeele T, Villunger P, Wagner AEF, et al. Essential versus accessory aspects of cell death: recommendations of the NCCD 2015. Cell Death Differ. 2015;22:58–73. - PMC - PubMed
-
- Buhl D, Snyder LE, Schwartz PR, Edrich J. HNCO in the Galactic Centre. Nature. 1973;243:513–514.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

