A-to-I editing of coding and non-coding RNAs by ADARs
- PMID: 26648264
- PMCID: PMC4824625
- DOI: 10.1038/nrm.2015.4
A-to-I editing of coding and non-coding RNAs by ADARs
Abstract
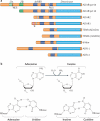
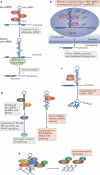
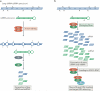
Adenosine deaminases acting on RNA (ADARs) convert adenosine to inosine in double-stranded RNA. This A-to-I editing occurs not only in protein-coding regions of mRNAs, but also frequently in non-coding regions that contain inverted Alu repeats. Editing of coding sequences can result in the expression of functionally altered proteins that are not encoded in the genome, whereas the significance of Alu editing remains largely unknown. Certain microRNA (miRNA) precursors are also edited, leading to reduced expression or altered function of mature miRNAs. Conversely, recent studies indicate that ADAR1 forms a complex with Dicer to promote miRNA processing, revealing a new function of ADAR1 in the regulation of RNA interference.
Figures


References
-
- Bass BL, Weintraub H. A developmentally regulated activity that unwinds RNA duplexes. Cell. 1987;48:607–613. - PubMed
-
- Bass BL, Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55:1089–1098. - PubMed
-
- Hogg M, Paro S, Keegan LP, O'Connell MA. RNA editing by mammalian ADARs. Adv. Genet. 2011;73:87–120. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

