Long-term neural and physiological phenotyping of a single human
- PMID: 26648521
- PMCID: PMC4682164
- DOI: 10.1038/ncomms9885
Long-term neural and physiological phenotyping of a single human
Abstract
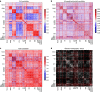
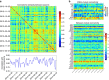
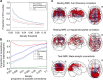
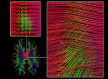
Psychiatric disorders are characterized by major fluctuations in psychological function over the course of weeks and months, but the dynamic characteristics of brain function over this timescale in healthy individuals are unknown. Here, as a proof of concept to address this question, we present the MyConnectome project. An intensive phenome-wide assessment of a single human was performed over a period of 18 months, including functional and structural brain connectivity using magnetic resonance imaging, psychological function and physical health, gene expression and metabolomics. A reproducible analysis workflow is provided, along with open access to the data and an online browser for results. We demonstrate dynamic changes in brain connectivity over the timescales of days to months, and relations between brain connectivity, gene expression and metabolites. This resource can serve as a testbed to study the joint dynamics of human brain and metabolic function over time, an approach that is critical for the development of precision medicine strategies for brain disorders.
Figures



References
Publication types
MeSH terms
Associated data
- Actions
- SRA/SRP042596
Grants and funding
- R01 NS046424/NS/NINDS NIH HHS/United States
- GM076536/GM/NIGMS NIH HHS/United States
- R01 GM076536/GM/NIGMS NIH HHS/United States
- DP1 GM106408/GM/NIGMS NIH HHS/United States
- OD009572/OD/NIH HHS/United States
- MH083824/MH/NIMH NIH HHS/United States
- F30 MH100872/MH/NIMH NIH HHS/United States
- R01 MH078111/MH/NIMH NIH HHS/United States
- MH078143/MH/NIMH NIH HHS/United States
- R24 HD042849/HD/NICHD NIH HHS/United States
- MH100872/MH/NIMH NIH HHS/United States
- R01 MH078143/MH/NIMH NIH HHS/United States
- NS0046424/NS/NINDS NIH HHS/United States
- R01 MH083824/MH/NIMH NIH HHS/United States
- 5R24HD042849/HD/NICHD NIH HHS/United States
- MH078111/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

