FOXM1 is a therapeutic target for high-risk multiple myeloma
- PMID: 26648534
- PMCID: PMC4826574
- DOI: 10.1038/leu.2015.334
FOXM1 is a therapeutic target for high-risk multiple myeloma
Abstract
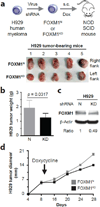
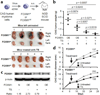
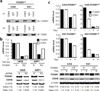
The transcription factor forkhead box M1 (FOXM1) is a validated oncoprotein in solid cancers, but its role in malignant plasma cell tumors such as multiple myeloma (MM) is unknown. We analyzed publicly available MM data sets and found that overexpression of FOXM1 prognosticates inferior outcome in a subset (~15%) of newly diagnosed cases, particularly patients with high-risk disease based on global gene expression changes. Follow-up studies using human myeloma cell lines (HMCLs) as the principal experimental model system demonstrated that enforced expression of FOXM1 increased growth, survival and clonogenicity of myeloma cells, whereas knockdown of FOXM1 abolished these features. In agreement with that, constitutive upregulation of FOXM1 promoted HMCL xenografts in laboratory mice, whereas inducible knockdown of FOXM1 led to growth inhibition. Expression of cyclin-dependent kinase 6 (CDK6) and NIMA-related kinase 2 (NEK2) was coregulated with FOXM1 in both HMCLs and myeloma patient samples, suggesting interaction of these three genes in a genetic network that may lend itself to targeting with small-drug inhibitors for new approaches to myeloma therapy and prevention. These results establish FOXM1 as high-risk myeloma gene and provide support for the design and testing of FOXM1-targeted therapies specifically for the FOXM1(High) subset of myeloma.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





References
-
- Shaughnessy JD, Jr, Zhan F, Burington BE, Huang Y, Colla S, Hanamura I, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007 Mar 15;109(6):2276–2284. - PubMed
-
- Kuiper R, Broyl A, de Knegt Y, van Vliet MH, van Beers EH, van der Holt B, et al. A gene expression signature for high-risk multiple myeloma. Leukemia. 2012 Nov;26(11):2406–2413. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

