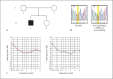
A Novel de novo Mutation in CEACAM16 Associated with Postlingual Hearing Impairment
- PMID: 26648831
- PMCID: PMC4662267
- DOI: 10.1159/000439576
A Novel de novo Mutation in CEACAM16 Associated with Postlingual Hearing Impairment
Abstract
Mutations in CEACAM16 cause autosomal dominant nonsyndromic hearing loss (DFNA4B). So far, 2 families have been reported with segregating missense mutations, both in the immunoglobulin constant domain A of the CEACAM16 protein. In this study, we used the TruSight One panel to investigate a parent-child trio without familial history of hearing loss and one affected child. When filtering for recessive inheritance and de novo events, we discovered a de novo CEACAM16 mutation (c.1094T>G, p.Leu365Arg) as the sole likely pathogenic variant. The de novo mutation was confirmed by Sanger sequencing and STR analysis. The proband's hearing loss closely matches the described onset and severity for DFNA4B. We present the third CEACAM16 variant and the first de novo mutation in CEACAM16. This de novo mutation is robustly described as a pathogenic mutation according to in silico mutation prediction tools and affects a highly conserved amino acid in the most strongly conserved CEACAM16 N2 domain. Our strategy of screening family trios enhances de novo mutation discovery and the exclusion of other variants of potential interest through pedigree filtering.
Keywords: Autosomal dominant nonsyndromic hearing loss; CEACAM16; DFNA4B; De novo mutation; Next-generation sequencing; Parent-child trios.
Figures

References
-
- Chen AH, Ni L, Fukushima K, Marietta J, O'Neill M, et al. Linkage of a gene for dominant non-syndromic deafness to chromosome 19. Hum Mol Genet. 1995;4:1073–1076. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

