Sequence-based Analysis of the Vitis vinifera L. cv Cabernet Sauvignon Grape Must Mycobiome in Three South African Vineyards Employing Distinct Agronomic Systems
- PMID: 26648930
- PMCID: PMC4663253
- DOI: 10.3389/fmicb.2015.01358
Sequence-based Analysis of the Vitis vinifera L. cv Cabernet Sauvignon Grape Must Mycobiome in Three South African Vineyards Employing Distinct Agronomic Systems
Abstract
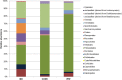
Recent microbiomic research of agricultural habitats has highlighted tremendous microbial biodiversity associated with such ecosystems. Data generated in vineyards have furthermore highlighted significant regional differences in vineyard biodiversity, hinting at the possibility that such differences might be responsible for regional differences in wine style and character, a hypothesis referred to as "microbial terroir." The current study further contributes to this body of work by comparing the mycobiome associated with South African (SA) Cabernet Sauvignon grapes in three neighboring vineyards that employ different agronomic approaches, and comparing the outcome with similar data sets from Californian vineyards. The aim of this study was to fully characterize the mycobiomes associated with the grapes from these vineyards. The data revealed approximately 10 times more fungal diversity than what is typically retrieved from culture-based studies. The Biodynamic vineyard was found to harbor a more diverse fungal community (H = 2.6) than the conventional (H = 2.1) and integrated (H = 1.8) vineyards. The data show that ascomycota are the most abundant phylum in the three vineyards, with Aureobasidium pullulans and its close relative Kabatiella microsticta being the most dominant fungi. This is the first report to reveal a high incidence of K. microsticta in the grape/wine ecosystem. Different common wine yeast species, such as Metschnikowia pulcherrima and Starmerella bacillaris dominated the mycobiome in the three vineyards. The data show that the filamentous fungi are the most abundant community in grape must although they are not regarded as relevant during wine fermentation. Comparison of metagenomic datasets from the three SA vineyards and previously published data from Californian vineyards revealed only 25% of the fungi in the SA dataset was also present in the Californian dataset, with greater variation evident amongst ubiquitous epiphytic fungi.
Keywords: grapevine mycobiome; microbial diversity; microbial terroir; next-generation sequencing; wine yeasts.
Figures






References
-
- Bagheri B., Bauer F. F., Setati M. E. (2015). The diversity and dynamics of indigenous yeast communities in grape must from vineyards employing different agronomic practices and their influence on wine fermentation. S. Afr. J. Enol. Vitic. 36 243–251.
LinkOut - more resources
Full Text Sources
Other Literature Sources

