One-Step Recovery of scFv Clones from High-Throughput Sequencing-Based Screening of Phage Display Libraries Challenged to Cells Expressing Native Claudin-1
- PMID: 26649313
- PMCID: PMC4662980
- DOI: 10.1155/2015/703213
One-Step Recovery of scFv Clones from High-Throughput Sequencing-Based Screening of Phage Display Libraries Challenged to Cells Expressing Native Claudin-1
Abstract
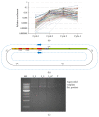
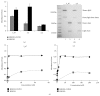
Expanding the availability of monoclonal antibodies interfering with hepatitis C virus infection of hepatocytes is an active field of investigation within medical biotechnologies, to prevent graft reinfection in patients subjected to liver transplantation and to overcome resistances elicited by novel antiviral drugs. In this paper, we describe a complete pipeline for screening of phage display libraries of human scFvs against native Claudin-1, a tight-junction protein involved in hepatitis C virus infection, expressed on the cell surface of human hepatocytes. To this aim, we implemented a high-throughput sequencing approach for library screening, followed by a simple and effective strategy to recover active binder clones from enriched sublibraries. The recovered clones were successfully converted to active immunoglobulins, thus demonstrating the effectiveness of the whole procedure. This novel approach can guarantee rapid and cheap isolation of antibodies for virtually any native antigen involved in human diseases, for therapeutic and/or diagnostic applications.
Figures

References
-
- Catanese M. T., Graziani R., von Hahn T., et al. High-avidity monoclonal antibodies against the human scavenger class B type I receptor efficiently block hepatitis C virus infection in the presence of high-density lipoprotein. Journal of Virology. 2007;81(15):8063–8071. doi: 10.1128/jvi.00193-07. - DOI - PMC - PubMed
-
- De Lorenzo C., Palmer D. B., Piccoli R., Ritter M. A., D'Alessio G. A. A new human antitumor immunoreagent specific for ErbB2. Clinical Cancer Research. 2002;8(6):1710–1719. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

