Newly Characterized Region of CP190 Associates with Microtubules and Mediates Proper Spindle Morphology in Drosophila Stem Cells
- PMID: 26649574
- PMCID: PMC4674064
- DOI: 10.1371/journal.pone.0144174
Newly Characterized Region of CP190 Associates with Microtubules and Mediates Proper Spindle Morphology in Drosophila Stem Cells
Abstract
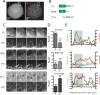
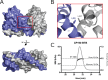
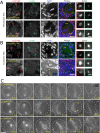
CP190 is a large, multi-domain protein, first identified as a centrosome protein with oscillatory localization over the course of the cell cycle. During interphase it has a well-established role within the nucleus as a chromatin insulator. Upon nuclear envelope breakdown, there is a striking redistribution of CP190 to centrosomes and the mitotic spindle, in addition to the population at chromosomes. Here, we investigate CP190 in detail by performing domain analysis in cultured Drosophila S2 cells combined with protein structure determination by X-ray crystallography, in vitro biochemical characterization, and in vivo fixed and live imaging of cp190 mutant flies. Our analysis of CP190 identifies a novel N-terminal centrosome and microtubule (MT) targeting region, sufficient for spindle localization. This region consists of a highly conserved BTB domain and a linker region that serves as the MT binding domain. We present the 2.5 Å resolution structure of the CP190 N-terminal 126 amino acids, which adopts a canonical BTB domain fold and exists as a stable dimer in solution. The ability of the linker region to robustly localize to MTs requires BTB domain-mediated dimerization. Deletion of the linker region using CRISPR significantly alters spindle morphology and leads to DNA segregation errors in the developing Drosophila brain neuroblasts. Collectively, we highlight a multivalent MT-binding architecture in CP190, which confers distinct subcellular cytoskeletal localization and function during mitosis.
Conflict of interest statement
Figures





Similar articles
-
The cell cycle-dependent localization of the CP190 centrosomal protein is determined by the coordinate action of two separable domains.J Cell Biol. 1995 Dec;131(5):1261-73. doi: 10.1083/jcb.131.5.1261. J Cell Biol. 1995. PMID: 8522588 Free PMC article.
-
The chromosomal association/dissociation of the chromatin insulator protein Cp190 of Drosophila melanogaster is mediated by the BTB/POZ domain and two acidic regions.BMC Cell Biol. 2010 Dec 31;11:101. doi: 10.1186/1471-2121-11-101. BMC Cell Biol. 2010. PMID: 21194420 Free PMC article.
-
The Drosophila centrosome-associated protein CP190 is essential for viability but not for cell division.J Cell Sci. 2004 Mar 1;117(Pt 7):1191-9. doi: 10.1242/jcs.00979. J Cell Sci. 2004. PMID: 14996941
-
Functional role of centrosomes in spindle assembly and organization.J Cell Biochem. 2004 Apr 1;91(5):904-14. doi: 10.1002/jcb.20013. J Cell Biochem. 2004. PMID: 15034926 Review.
-
Cell division: MAST sails through mitosis.Curr Biol. 2002 Sep 3;12(17):R585-7. doi: 10.1016/s0960-9822(02)01098-9. Curr Biol. 2002. PMID: 12225678 Review.
Cited by
-
Multiple Roles of dXNP and dADD1-Drosophila Orthologs of ATRX Chromatin Remodeler.Int J Mol Sci. 2023 Nov 18;24(22):16486. doi: 10.3390/ijms242216486. Int J Mol Sci. 2023. PMID: 38003676 Free PMC article. Review.
-
The N-Terminal Part of Drosophila CP190 Is a Platform for Interaction with Multiple Architectural Proteins.Int J Mol Sci. 2023 Nov 2;24(21):15917. doi: 10.3390/ijms242115917. Int J Mol Sci. 2023. PMID: 37958900 Free PMC article.
-
HIPP1 stabilizes the interaction between CP190 and Su(Hw) in the Drosophila insulator complex.Sci Rep. 2019 Dec 13;9(1):19102. doi: 10.1038/s41598-019-55617-6. Sci Rep. 2019. PMID: 31836797 Free PMC article.
-
Mechanisms of CP190 Interaction with Architectural Proteins in Drosophila Melanogaster.Int J Mol Sci. 2021 Nov 17;22(22):12400. doi: 10.3390/ijms222212400. Int J Mol Sci. 2021. PMID: 34830280 Free PMC article.
-
Role of Su(Hw) zinc finger 10 and interaction with CP190 and Mod(mdg4) proteins in recruiting the Su(Hw) complex to chromatin sites in Drosophila.PLoS One. 2018 Feb 23;13(2):e0193497. doi: 10.1371/journal.pone.0193497. eCollection 2018. PLoS One. 2018. PMID: 29474480 Free PMC article.
References
-
- Mitchison T, Kirschner M (1984) Dynamic instability of microtubule growth. Nature 312: 237–242. - PubMed
-
- Weisenbe Rc (1972) Microtubule Formation in-Vitro in Solutions Containing Low Calcium Concentrations. Science 177: 1104–&. - PubMed
-
- Moritz M, Braunfeld MB, Sedat JW, Alberts B, Agard DA (1995) Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature 378: 638–640. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

