Structure and functional characterization of a bile acid 7α dehydratase BaiE in secondary bile acid synthesis
- PMID: 26650892
- PMCID: PMC4755848
- DOI: 10.1002/prot.24971
Structure and functional characterization of a bile acid 7α dehydratase BaiE in secondary bile acid synthesis
Abstract
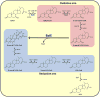
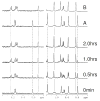
Conversion of the primary bile acids cholic acid (CA) and chenodeoxycholic acid (CDCA) to the secondary bile acids deoxycholic acid (DCA) and lithocholic acid (LCA) is performed by a few species of intestinal bacteria in the genus Clostridium through a multistep biochemical pathway that removes a 7α-hydroxyl group. The rate-determining enzyme in this pathway is bile acid 7α-dehydratase (baiE). In this study, crystal structures of apo-BaiE and its putative product-bound [3-oxo-Δ(4,6) -lithocholyl-Coenzyme A (CoA)] complex are reported. BaiE is a trimer with a twisted α + β barrel fold with similarity to the Nuclear Transport Factor 2 (NTF2) superfamily. Tyr30, Asp35, and His83 form a catalytic triad that is conserved across this family. Site-directed mutagenesis of BaiE from Clostridium scindens VPI 12708 confirm that these residues are essential for catalysis and also the importance of other conserved residues, Tyr54 and Arg146, which are involved in substrate binding and affect catalytic turnover. Steady-state kinetic studies reveal that the BaiE homologs are able to turn over 3-oxo-Δ(4) -bile acid and CoA-conjugated 3-oxo-Δ(4) -bile acid substrates with comparable efficiency questioning the role of CoA-conjugation in the bile acid metabolism pathway.
Keywords: 7α-dehyroxylation; bile acid 7α-dehydratase; gut microbe mediated human metabolite; gut microbes; nuclear transport factor-2 superfamily; primary bile acid; secondary bile acid; secondary bile acid synthesis; structural genomics.
© 2015 Wiley Periodicals, Inc.
Figures

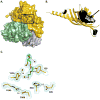



Similar articles
-
Clostridium scindens ATCC 35704: Integration of Nutritional Requirements, the Complete Genome Sequence, and Global Transcriptional Responses to Bile Acids.Appl Environ Microbiol. 2019 Mar 22;85(7):e00052-19. doi: 10.1128/AEM.00052-19. Print 2019 Apr 1. Appl Environ Microbiol. 2019. PMID: 30737348 Free PMC article.
-
Identification and characterization of a bile acid 7alpha-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7alpha-dehydroxylating strain isolated from human feces.Appl Environ Microbiol. 2000 Mar;66(3):1107-13. doi: 10.1128/AEM.66.3.1107-1113.2000. Appl Environ Microbiol. 2000. PMID: 10698778 Free PMC article.
-
Metabolism of Oxo-Bile Acids and Characterization of Recombinant 12α-Hydroxysteroid Dehydrogenases from Bile Acid 7α-Dehydroxylating Human Gut Bacteria.Appl Environ Microbiol. 2018 May 1;84(10):e00235-18. doi: 10.1128/AEM.00235-18. Print 2018 May 15. Appl Environ Microbiol. 2018. PMID: 29549099 Free PMC article.
-
Identification and characterization of two bile acid coenzyme A transferases from Clostridium scindens, a bile acid 7α-dehydroxylating intestinal bacterium.J Lipid Res. 2012 Jan;53(1):66-76. doi: 10.1194/jlr.M020313. Epub 2011 Oct 20. J Lipid Res. 2012. PMID: 22021638 Free PMC article.
-
Structural and functional characterization of BaiA, an enzyme involved in secondary bile acid synthesis in human gut microbe.Proteins. 2014 Feb;82(2):216-29. doi: 10.1002/prot.24353. Epub 2013 Oct 17. Proteins. 2014. PMID: 23836456 Free PMC article.
Cited by
-
7β-Hydroxysteroid dehydratase Hsh3 eliminates the 7-hydroxy group of the bile salt ursodeoxycholate during degradation by Sphingobium sp. strain Chol11 and other Sphingomonadaceae.Appl Environ Microbiol. 2025 Jun 18;91(6):e0018525. doi: 10.1128/aem.00185-25. Epub 2025 May 9. Appl Environ Microbiol. 2025. PMID: 40340444 Free PMC article.
-
Integrative analysis of metabolome and gut microbiota in diet-induced hyperlipidemic rats treated with berberine compounds.J Transl Med. 2016 Aug 5;14(1):237. doi: 10.1186/s12967-016-0987-5. J Transl Med. 2016. PMID: 27495782 Free PMC article.
-
The 7-α-dehydroxylation pathway: An integral component of gut bacterial bile acid metabolism and potential therapeutic target.Front Microbiol. 2023 Jan 9;13:1093420. doi: 10.3389/fmicb.2022.1093420. eCollection 2022. Front Microbiol. 2023. PMID: 36699589 Free PMC article. Review.
-
Formation of secondary allo-bile acids by novel enzymes from gut Firmicutes.Gut Microbes. 2022 Jan-Dec;14(1):2132903. doi: 10.1080/19490976.2022.2132903. Gut Microbes. 2022. PMID: 36343662 Free PMC article.
-
Clostridium scindens ATCC 35704: Integration of Nutritional Requirements, the Complete Genome Sequence, and Global Transcriptional Responses to Bile Acids.Appl Environ Microbiol. 2019 Mar 22;85(7):e00052-19. doi: 10.1128/AEM.00052-19. Print 2019 Apr 1. Appl Environ Microbiol. 2019. PMID: 30737348 Free PMC article.
References
-
- Sonnenburg JL, Xu J, Leip DD, Chen C-H, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. - PubMed
-
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. - PubMed
-
- Lupton JR. Microbial degradation products influence colon cancer risk: the butyrate controversy. J Nutr. 2004;134:479–482. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

