Improved taxonomic assignment of human intestinal 16S rRNA sequences by a dedicated reference database
- PMID: 26651617
- PMCID: PMC4676846
- DOI: 10.1186/s12864-015-2265-y
Improved taxonomic assignment of human intestinal 16S rRNA sequences by a dedicated reference database
Abstract
Background: Current sequencing technology enables taxonomic profiling of microbial ecosystems at high resolution and depth by using the 16S rRNA gene as a phylogenetic marker. Taxonomic assignation of newly acquired data is based on sequence comparisons with comprehensive reference databases to find consensus taxonomy for representative sequences. Nevertheless, even with well-characterised ecosystems like the human intestinal microbiota it is challenging to assign genus and species level taxonomy to 16S rRNA amplicon reads. A part of the explanation may lie in the sheer size of the search space where competition from a multitude of highly similar sequences may not allow reliable assignation at low taxonomic levels. However, when studying a particular environment such as the human intestine, it can be argued that a reference database comprising only sequences that are native to the environment would be sufficient, effectively reducing the search space.
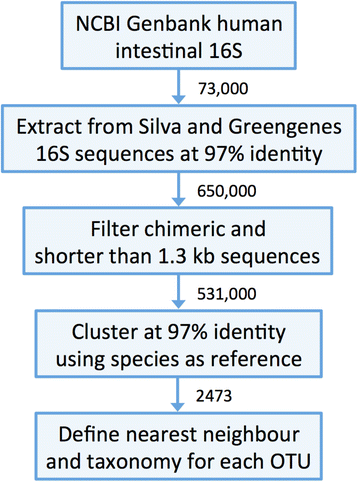
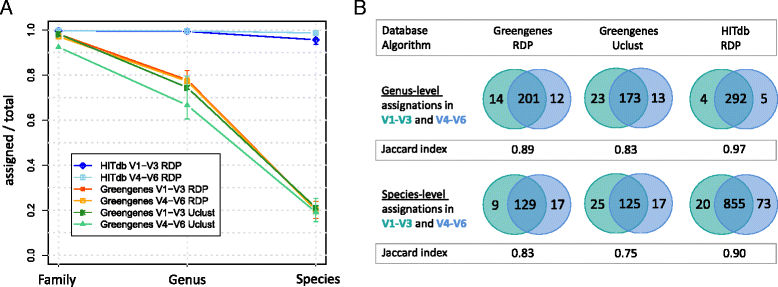
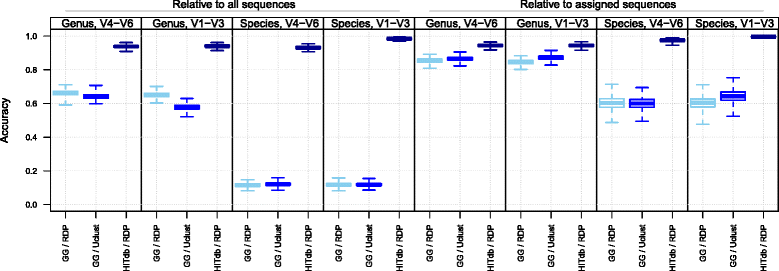
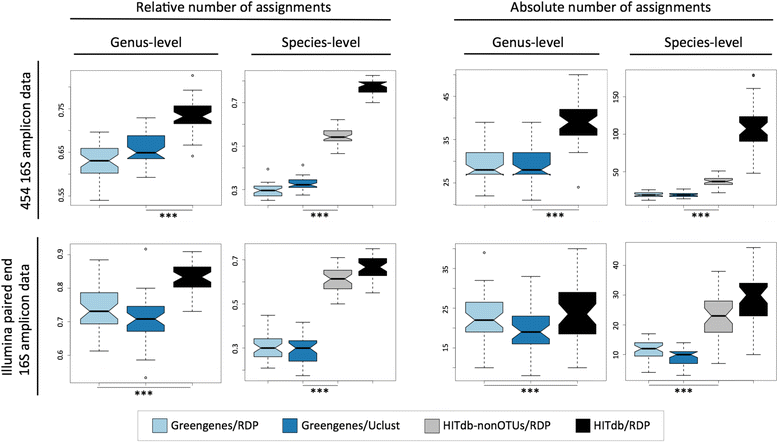
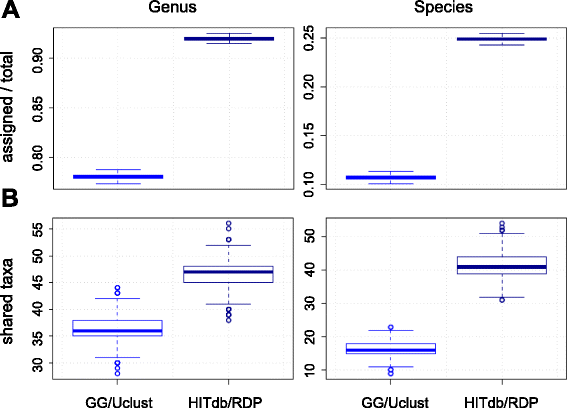
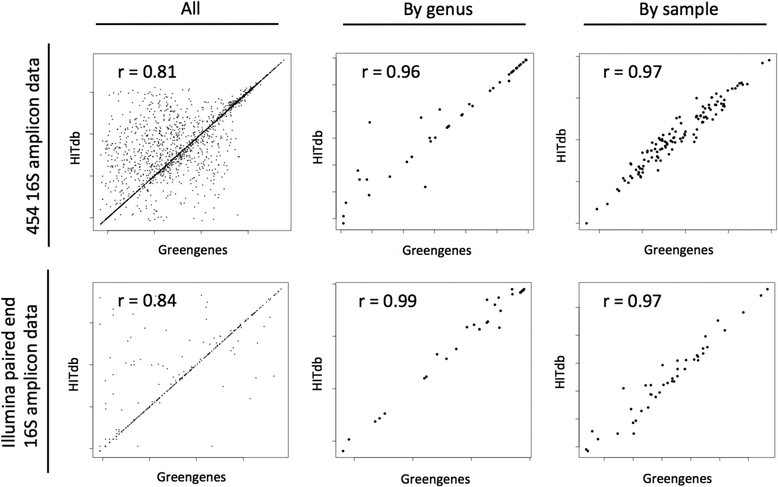
Results: We constructed a 16S rRNA gene database based on high-quality sequences specific for human intestinal microbiota, resulting in curated data set consisting of 2473 unique prokaryotic species-like groups and their taxonomic lineages, and compared its performance against the Greengenes and Silva databases. The results showed that regardless of used assignment algorithm, our database improved taxonomic assignation of 16S rRNA sequencing data by enabling significantly higher species and genus level assignation rate while preserving taxonomic diversity and demanding less computational resources.
Conclusion: The curated human intestinal 16S rRNA gene taxonomic database of about 2500 species-like groups described here provides a practical solution for significantly improved taxonomic assignment for phylogenetic studies of the human intestinal microbiota.
Figures
Similar articles
-
TaxAss: Leveraging a Custom Freshwater Database Achieves Fine-Scale Taxonomic Resolution.mSphere. 2018 Sep 5;3(5):e00327-18. doi: 10.1128/mSphere.00327-18. mSphere. 2018. PMID: 30185512 Free PMC article.
-
Construction & assessment of a unified curated reference database for improving the taxonomic classification of bacteria using 16S rRNA sequence data.Indian J Med Res. 2020 Jan;151(1):93-103. doi: 10.4103/ijmr.IJMR_220_18. Indian J Med Res. 2020. PMID: 32134020 Free PMC article.
-
Construction of habitat-specific training sets to achieve species-level assignment in 16S rRNA gene datasets.Microbiome. 2020 May 15;8(1):65. doi: 10.1186/s40168-020-00841-w. Microbiome. 2020. PMID: 32414415 Free PMC article.
-
GSR-DB: a manually curated and optimized taxonomical database for 16S rRNA amplicon analysis.mSystems. 2024 Feb 20;9(2):e0095023. doi: 10.1128/msystems.00950-23. Epub 2024 Jan 8. mSystems. 2024. PMID: 38189256 Free PMC article.
-
A step-by-step procedure for analysing the 16S rRNA-based microbiome diversity using QIIME 2 and comprehensive PICRUSt2 illustration for functional prediction.Arch Microbiol. 2024 Nov 14;206(12):467. doi: 10.1007/s00203-024-04177-z. Arch Microbiol. 2024. PMID: 39540937 Review.
Cited by
-
The Influences of Bioinformatics Tools and Reference Databases in Analyzing the Human Oral Microbial Community.Genes (Basel). 2020 Aug 3;11(8):878. doi: 10.3390/genes11080878. Genes (Basel). 2020. PMID: 32756341 Free PMC article.
-
The Obesity-Related Gut Bacterial and Viral Dysbiosis Can Impact the Risk of Colon Cancer Development.Microorganisms. 2020 Mar 19;8(3):431. doi: 10.3390/microorganisms8030431. Microorganisms. 2020. PMID: 32204328 Free PMC article.
-
Host dispersal relaxes selective pressures in rafting microbiomes and triggers successional changes.Nat Commun. 2024 Dec 30;15(1):10759. doi: 10.1038/s41467-024-54954-z. Nat Commun. 2024. PMID: 39737966 Free PMC article.
-
CoMA - an intuitive and user-friendly pipeline for amplicon-sequencing data analysis.PLoS One. 2020 Dec 2;15(12):e0243241. doi: 10.1371/journal.pone.0243241. eCollection 2020. PLoS One. 2020. PMID: 33264369 Free PMC article.
-
Characterization of the Gut Microbiome Using 16S or Shotgun Metagenomics.Front Microbiol. 2016 Apr 20;7:459. doi: 10.3389/fmicb.2016.00459. eCollection 2016. Front Microbiol. 2016. PMID: 27148170 Free PMC article.
References
-
- Cheng J, Palva AM, de Vos WM, Satokari R. Contribution of the intestinal microbiota to human health: from birth to 100 years of age. Curr Top Microbiol Immunol. 2013;358:323–346. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

