Population genomics of intrapatient HIV-1 evolution
- PMID: 26652000
- PMCID: PMC4718817
- DOI: 10.7554/eLife.11282
Population genomics of intrapatient HIV-1 evolution
Abstract
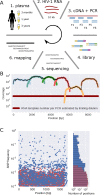
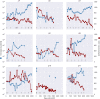
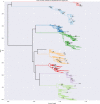
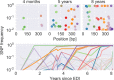
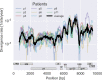
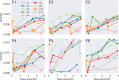
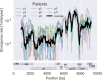
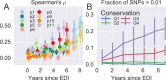
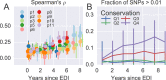
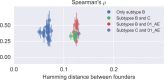
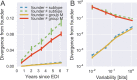
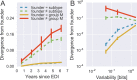
Many microbial populations rapidly adapt to changing environments with multiple variants competing for survival. To quantify such complex evolutionary dynamics in vivo, time resolved and genome wide data including rare variants are essential. We performed whole-genome deep sequencing of HIV-1 populations in 9 untreated patients, with 6-12 longitudinal samples per patient spanning 5-8 years of infection. The data can be accessed and explored via an interactive web application. We show that patterns of minor diversity are reproducible between patients and mirror global HIV-1 diversity, suggesting a universal landscape of fitness costs that control diversity. Reversions towards the ancestral HIV-1 sequence are observed throughout infection and account for almost one third of all sequence changes. Reversion rates depend strongly on conservation. Frequent recombination limits linkage disequilibrium to about 100 bp in most of the genome, but strong hitch-hiking due to short range linkage limits diversity.
Keywords: HIV-1; deep sequencing; evolution; evolutionary biology; fitness landscapes; genomics; infectious disease; microbiology; reversion; viruses.
Conflict of interest statement
RAN: Reviewing editor,
The other authors declare that no competing interests exist.
Figures

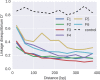

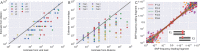

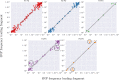
References
-
- Allen TM, Altfeld M, Geer SC, Kalife ET, Moore C, O'Sullivan KM, DeSouza I, Feeney ME, Eldridge RL, Maier EL, Kaufmann DE, Lahaie MP, Reyor L, Tanzi G, Johnston MN, Brander C, Draenert R, Rockstroh JK, Jessen H, Rosenberg ES, Mallal SA, Walker BD. Selective escape from CD8+ t-cell responses represents a major driving force of human immunodeficiency virus type 1 (hIV-1) sequence diversity and reveals constraints on HIV-1 evolution. Journal of Virology. 2005;79:13239–13249. doi: 10.1128/JVI.79.21.13239-13249.2005. - DOI - PMC - PubMed
-
- Bar KJ, Tsao Chun-yen, Iyer SS, Decker JM, Yang Y, Bonsignori M, Chen X, Hwang K-K, Montefiori DC, Liao H-X, Hraber P, Fischer W, Li H, Wang S, Sterrett S, Keele BF, Ganusov VV, Perelson AS, Korber BT, Georgiev I, McLellan JS, Pavlicek JW, Gao F, Haynes BF, Hahn BH, Kwong PD, Shaw GM, Trkola A. Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape. PLoS Pathogens. 2012;8:e11282. doi: 10.1371/journal.ppat.1002721. - DOI - PMC - PubMed
-
- Batorsky R, Kearney MF, Palmer SE, Maldarelli F, Rouzine IM, Coffin JM. Estimate of effective recombination rate and average selection coefficient for HIV in chronic infection. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5661–5666. doi: 10.1073/pnas.1102036108. - DOI - PMC - PubMed
-
- Bernardin F, Kong D, Peddada L, Baxter-Lowe LA, Delwart E. Human immunodeficiency virus mutations during the first month of infection are preferentially found in known cytotoxic t-lymphocyte epitopes. Journal of Virology. 2005;79:11523–11528. doi: 10.1128/JVI.79.17.11523-11528.2005. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

