Understanding differences between synthetic and natural antibodies can help improve antibody engineering
- PMID: 26652053
- PMCID: PMC4966605
- DOI: 10.1080/19420862.2015.1123365
Understanding differences between synthetic and natural antibodies can help improve antibody engineering
Abstract
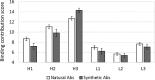
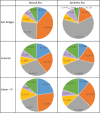
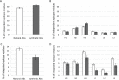
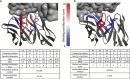
Synthetic libraries are a major source of human-like antibody (Ab) drug leads. To assess the similarity between natural Abs and the products of these libraries, we compared large sets of natural and synthetic Abs using "CDRs Analyzer," a tool we introduce for structural analysis of Ab-antigen (Ag) interactions. Natural Abs, we found, recognize their Ags by combining multiple complementarity-determining regions (CDRs) to create an integrated interface. Synthetic Abs, however, rely dominantly, sometimes even exclusively on CDRH3. The increased contribution of CDRH3 to Ag binding in synthetic Abs comes with a substantial decrease in the involvement of CDRH2 and CDRH1. Furthermore, in natural Abs CDRs specialize in specific types of non-covalent interactions with the Ag. CDRH1 accounts for a significant portion of the cation-pi interactions; CDRH2 is the major source of salt-bridges and CDRH3 accounts for most hydrogen bonds. In synthetic Abs this specialization is lost, and CDRH3 becomes the main sources of all types of contacts. The reliance of synthetic Abs on CDRH3 reduces the complexity of their interaction with the Ag: More Ag residues contact only one CDR and fewer contact 3 CDRs or more. We suggest that the focus of engineering attempts on CDRH3 results in libraries enriched with variants that are not natural-like. This may affect not only Ag binding, but also Ab expression, stability and selectivity. Our findings can help guide library design, creating libraries that can bind more epitopes and Abs that better mimic the natural antigenic interactions.
Keywords: Antigen binding site; antibody–antigen interactions; human-like antibodies; libraries; synthetic.
Figures
References
-
- Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975; 256:495-7; PMID:1172191; http://dx.doi.org/10.1038/256495a0. - DOI - PubMed
-
- Kung P, Goldstein G, Reinherz EL, Schlossman SF. Monoclonal antibodies defining distinctive human T cell surface antigens. Science 1979; 206:347-9; PMID:314668; http://dx.doi.org/10.1126/science.314668. - DOI - PubMed
-
- Kinch MS. An overview of FDA-approved biologics medicines. Drug Discov Today 2015; 20:393-8; PMID:25220442; http://dx.doi.org/10.1016/j.drudis.2014.09.003. - DOI - PubMed
-
- Janeway CA Jr, Travers P, Walport M, Shlomchik MJ Immunobiology: The Immune System in Health and Disease. 5th edition. New York: Garland Science; 2001.
-
- Novotný J, Handschumacher M, Haber E, Bruccoleri RE, Carlson WB, Fanning DW, Smith JA, Rose GD. Antigenic determinants in proteins coincide with surface regions accessible to large probes (antibody domains). Proc Natl Acad Sci U S A 1986; 83:226-30; PMID:2417241; http://dx.doi.org/10.1073/pnas.83.2.226. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
