Coordination of DNA damage tolerance mechanisms with cell cycle progression in fission yeast
- PMID: 26652183
- PMCID: PMC5007584
- DOI: 10.1080/15384101.2015.1121353
Coordination of DNA damage tolerance mechanisms with cell cycle progression in fission yeast
Abstract
DNA damage tolerance (DDT) mechanisms allow cells to synthesize a new DNA strand when the template is damaged. Many mutations resulting from DNA damage in eukaryotes are generated during DDT when cells use the mutagenic translesion polymerases, Rev1 and Polζ, rather than mechanisms with higher fidelity. The coordination among DDT mechanisms is not well understood. We used live-cell imaging to study the function of DDT mechanisms throughout the cell cycle of the fission yeast Schizosaccharomyces pombe. We report that checkpoint-dependent mitotic delay provides a cellular mechanism to ensure the completion of high fidelity DDT, largely by homology-directed repair (HDR). DDT by mutagenic polymerases is suppressed during the checkpoint delay by a mechanism dependent on Rad51 recombinase. When cells pass the G2/M checkpoint and can no longer delay mitosis, they completely lose the capacity for HDR and simultaneously exhibit a requirement for Rev1 and Polζ. Thus, DDT is coordinated with the checkpoint response so that the activity of mutagenic polymerases is confined to a vulnerable period of the cell cycle when checkpoint delay and HDR are not possible.
Keywords: Chk1; DNA damage checkpoint; DNA damage tolerance; Polζ; Polη; Rev1; homology-directed repair; post-replication repair; ubiquitin ligase Rad5.
Figures



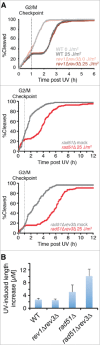

Comment in
-
Non mutagenic and mutagenic DNA damage tolerance.Cell Cycle. 2016;15(3):314-5. doi: 10.1080/15384101.2015.1132909. Cell Cycle. 2016. PMID: 26727652 Free PMC article. No abstract available.
Similar articles
-
Postreplication gaps at UV lesions are signals for checkpoint activation.Proc Natl Acad Sci U S A. 2010 May 4;107(18):8219-24. doi: 10.1073/pnas.1003449107. Epub 2010 Apr 19. Proc Natl Acad Sci U S A. 2010. PMID: 20404181 Free PMC article.
-
The Protein Level of Rev1, a TLS Polymerase in Fission Yeast, Is Strictly Regulated during the Cell Cycle and after DNA Damage.PLoS One. 2015 Jul 6;10(7):e0130000. doi: 10.1371/journal.pone.0130000. eCollection 2015. PLoS One. 2015. PMID: 26147350 Free PMC article.
-
Lingering single-strand breaks trigger Rad51-independent homology-directed repair of collapsed replication forks in the polynucleotide kinase/phosphatase mutant of fission yeast.PLoS Genet. 2017 Sep 18;13(9):e1007013. doi: 10.1371/journal.pgen.1007013. eCollection 2017 Sep. PLoS Genet. 2017. PMID: 28922417 Free PMC article.
-
All eukaryotes: before turning off G1-S transcription, please check your DNA.Cell Cycle. 2009 Jan 15;8(2):214-7. doi: 10.4161/cc.8.2.7412. Cell Cycle. 2009. PMID: 19158488 Review.
-
The roles of DNA polymerase ζ and the Y family DNA polymerases in promoting or preventing genome instability.Mutat Res. 2013 Mar-Apr;743-744:97-110. doi: 10.1016/j.mrfmmm.2012.11.002. Epub 2012 Nov 26. Mutat Res. 2013. PMID: 23195997 Free PMC article. Review.
Cited by
-
Solution NMR structure of the HLTF HIRAN domain: a conserved module in SWI2/SNF2 DNA damage tolerance proteins.J Biomol NMR. 2016 Nov;66(3):209-219. doi: 10.1007/s10858-016-0070-9. Epub 2016 Oct 22. J Biomol NMR. 2016. PMID: 27771863
-
Mathematical description of eukaryotic chromosome replication.Proc Natl Acad Sci U S A. 2019 Mar 12;116(11):4776-4778. doi: 10.1073/pnas.1900968116. Epub 2019 Feb 19. Proc Natl Acad Sci U S A. 2019. PMID: 30782813 Free PMC article. No abstract available.
-
Translesion synthesis polymerases contribute to meiotic chromosome segregation and cohesin dynamics in Schizosaccharomycespombe.J Cell Sci. 2020 May 22;133(10):jcs238709. doi: 10.1242/jcs.238709. J Cell Sci. 2020. PMID: 32317395 Free PMC article.
-
Dynamics of DNA replication in a eukaryotic cell.Proc Natl Acad Sci U S A. 2019 Mar 12;116(11):4973-4982. doi: 10.1073/pnas.1818680116. Epub 2019 Feb 4. Proc Natl Acad Sci U S A. 2019. PMID: 30718387 Free PMC article.
-
Non mutagenic and mutagenic DNA damage tolerance.Cell Cycle. 2016;15(3):314-5. doi: 10.1080/15384101.2015.1132909. Cell Cycle. 2016. PMID: 26727652 Free PMC article. No abstract available.
References
-
- Brash DE. UV signature mutations. Photochem Photobiol 2015; 91:15–26; PMID:25354245; http://dx.doi.org/10.1111/php.12377 - DOI - PMC - PubMed
-
- Callegari AJ, Kelly TJ. Shedding light on the DNA damage checkpoint. Cell Cycle 2007; 6:660–6; PMID:17387276; http://dx.doi.org/10.4161/cc.6.6.3984 - DOI - PubMed
-
- Lemontt JF. Genetic and physiological factors affecting repair and mutagenesis in yeast. Basic Life Sci 1980; 15:85–120; PMID:7011312 - PubMed
-
- Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev 2009; 73:134–54; PMID:19258535; http://dx.doi.org/10.1128/MMBR.00034-08 - DOI - PMC - PubMed
-
- Sale JE. Competition, collaboration and coordination–determining how cells bypass DNA damage. J Cell Sci 2012; 125:1633–43; PMID:22499669; http://dx.doi.org/10.1242/jcs.094748 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
