Combined de novo and genome guided assembly and annotation of the Pinus patula juvenile shoot transcriptome
- PMID: 26652261
- PMCID: PMC4676862
- DOI: 10.1186/s12864-015-2277-7
Combined de novo and genome guided assembly and annotation of the Pinus patula juvenile shoot transcriptome
Abstract
Background: Pines are the most important tree species to the international forestry industry, covering 42 % of the global industrial forest plantation area. One of the most pressing threats to cultivation of some pine species is the pitch canker fungus, Fusarium circinatum, which can have devastating effects in both the field and nursery. Investigation of the Pinus-F. circinatum host-pathogen interaction is crucial for development of effective disease management strategies. As with many non-model organisms, investigation of host-pathogen interactions in pine species is hampered by limited genomic resources. This was partially alleviated through release of the 22 Gbp Pinus taeda v1.01 genome sequence ( http://pinegenome.org/pinerefseq/ ) in 2014. Despite the fact that the fragmented state of the genome may hamper comprehensive transcriptome analysis, it is possible to leverage the inherent redundancy resulting from deep RNA sequencing with Illumina short reads to assemble transcripts in the absence of a completed reference sequence. These data can then be integrated with available genomic data to produce a comprehensive transcriptome resource. The aim of this study was to provide a foundation for gene expression analysis of disease response mechanisms in Pinus patula through transcriptome assembly.
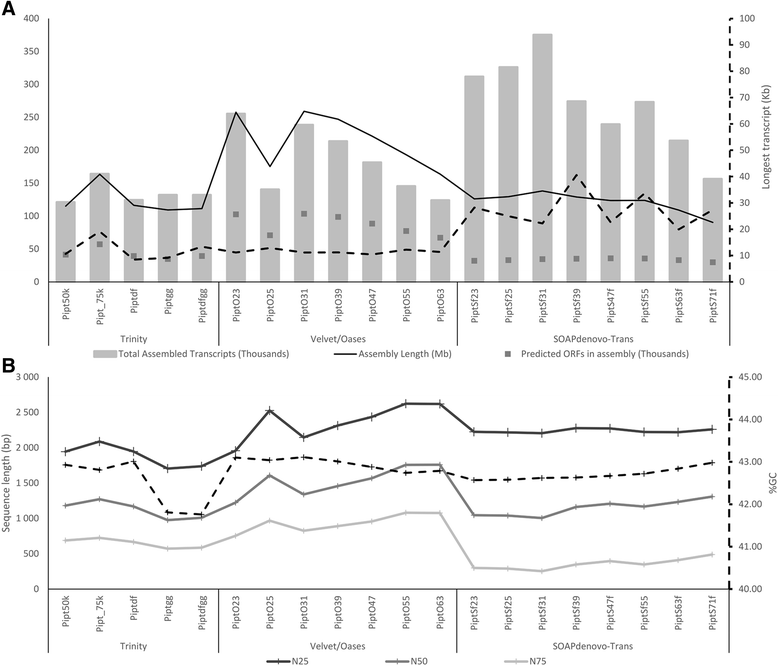
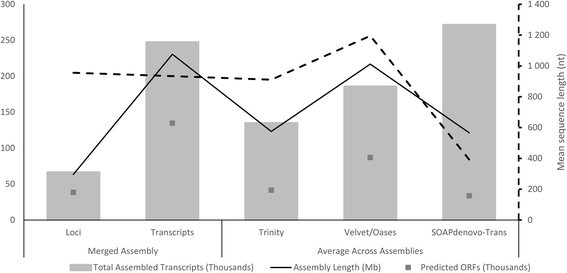
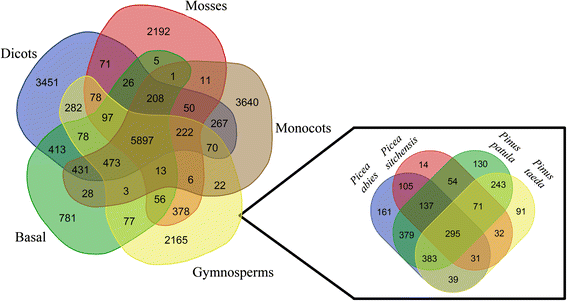
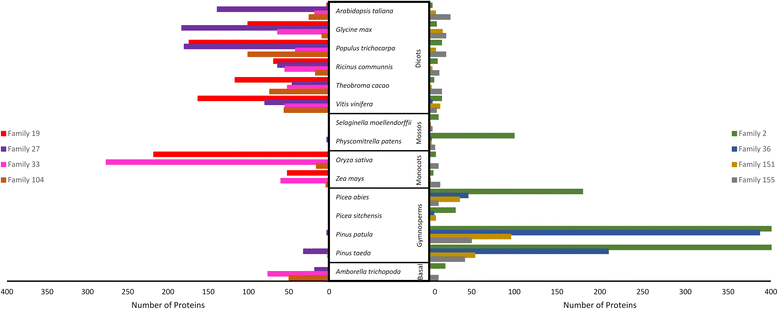
Results: Eighteen de novo and two reference based assemblies were produced for P. patula shoot tissue. For this purpose three transcriptome assemblers, Trinity, Velvet/OASES and SOAPdenovo-Trans, were used to maximise diversity and completeness of assembled transcripts. Redundancy in the assembly was reduced using the EvidentialGene pipeline. The resulting 52 Mb P. patula v1.0 shoot transcriptome consists of 52 112 unigenes, 60 % of which could be functionally annotated.
Conclusions: The assembled transcriptome will serve as a major genomic resource for future investigation of P. patula and represents the largest gene catalogue produced to date for this species. Furthermore, this assembly can help detect gene-based genetic markers for P. patula and the comparative assembly workflow could be applied to generate similar resources for other non-model species.
Figures
Similar articles
-
Defence transcriptome assembly and pathogenesis related gene family analysis in Pinus tecunumanii (low elevation).BMC Genomics. 2018 Aug 23;19(1):632. doi: 10.1186/s12864-018-5015-0. BMC Genomics. 2018. PMID: 30139335 Free PMC article.
-
Transcriptome sequencing in an ecologically important tree species: assembly, annotation, and marker discovery.BMC Genomics. 2010 Mar 16;11:180. doi: 10.1186/1471-2164-11-180. BMC Genomics. 2010. PMID: 20233449 Free PMC article.
-
De novo assembly of maritime pine transcriptome: implications for forest breeding and biotechnology.Plant Biotechnol J. 2014 Apr;12(3):286-99. doi: 10.1111/pbi.12136. Epub 2013 Nov 21. Plant Biotechnol J. 2014. PMID: 24256179
-
Strategies for transcriptome analysis in nonmodel plants.Am J Bot. 2012 Feb;99(2):267-76. doi: 10.3732/ajb.1100334. Epub 2012 Feb 1. Am J Bot. 2012. PMID: 22301897 Review.
-
[Transcript assembly and quality assessment].Sheng Wu Gong Cheng Xue Bao. 2015 Sep;31(9):1271-8. Sheng Wu Gong Cheng Xue Bao. 2015. PMID: 26955705 Review. Chinese.
Cited by
-
Assessing the Gene Content of the Megagenome: Sugar Pine (Pinus lambertiana).G3 (Bethesda). 2016 Dec 7;6(12):3787-3802. doi: 10.1534/g3.116.032805. G3 (Bethesda). 2016. PMID: 27799338 Free PMC article.
-
Transcriptome sequencing reveals signatures of positive selection in the Spot-Tailed Earless Lizard.PLoS One. 2020 Jun 15;15(6):e0234504. doi: 10.1371/journal.pone.0234504. eCollection 2020. PLoS One. 2020. PMID: 32542006 Free PMC article.
-
Comprehensive assembly and analysis of the transcriptome of maritime pine developing embryos.BMC Plant Biol. 2018 Dec 29;18(1):379. doi: 10.1186/s12870-018-1564-2. BMC Plant Biol. 2018. PMID: 30594130 Free PMC article.
-
The transcriptome of Pinus pinaster under Fusarium circinatum challenge.BMC Genomics. 2020 Jan 8;21(1):28. doi: 10.1186/s12864-019-6444-0. BMC Genomics. 2020. PMID: 31914917 Free PMC article.
-
Elucidating the Mesocarp Drupe Transcriptome of Açai (Euterpe oleracea Mart.): An Amazonian Tree Palm Producer of Bioactive Compounds.Int J Mol Sci. 2023 May 26;24(11):9315. doi: 10.3390/ijms24119315. Int J Mol Sci. 2023. PMID: 37298279 Free PMC article.
References
-
- Critchfield W, Little E. Geographic distribution of pines of the world. USDA For Serv. 1966;991:1–97.
-
- Indufor: Forest Stewardship Council (FSC) Strategic Review on the Future of Forest Plantations. 2012:121.
-
- Wingfield MJ, Coutinho TA, Roux J, Wingfield BD. The future of exotic plantation forestry in the tropics and southern Hemisphere: Lessons from pitch canker. South Afr Forestry J. 2002;195:79–82. doi: 10.1080/20702620.2002.10434607. - DOI
-
- Wingfield MJ, Hammerbacher A, Ganley RJ, Steenkamp ET, Gordon TR, Wingfield BD, et al. Pitch canker caused by Fusarium circinatum - A growing threat to pine plantations and forests worldwide. Australas Plant Pathol. 2008;37:319–334. doi: 10.1071/AP08036. - DOI
-
- Hodge GR, Dvorak WS. Differential responses of Central American and Mexican pine species and Pinus radiata to infection by the pitch canker fungus. New For. 2000;19:241–258. doi: 10.1023/A:1006613021996. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

