Relative and contextual contribution of different sources to the composition and abundance of indoor air bacteria in residences
- PMID: 26653310
- PMCID: PMC4674937
- DOI: 10.1186/s40168-015-0128-z
Relative and contextual contribution of different sources to the composition and abundance of indoor air bacteria in residences
Abstract
Background: The study of the microbial communities in the built environment is of critical importance as humans spend the majority of their time indoors. While the microorganisms in living spaces, especially those in the air, can impact health and well-being, little is known of their identity and the processes that determine their assembly. We investigated the source-sink relationships of airborne bacteria in 29 homes in the San Francisco Bay Area. Samples taken in the sites expected to be source habitats for indoor air microbes were analyzed by 16S rRNA-based pyrosequencing and quantitative PCR. The community composition was related to the characteristics of the household collected at the time of sampling, including the number of residents and pets, activity levels, frequency of cooking and vacuum cleaning, extent of natural ventilation, and abundance and type of vegetation surrounding the building.
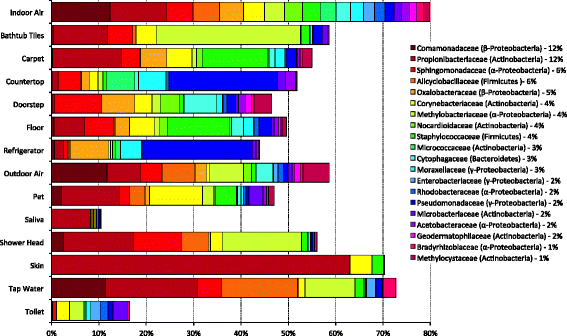
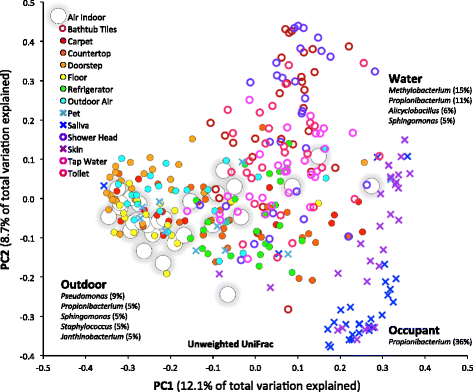
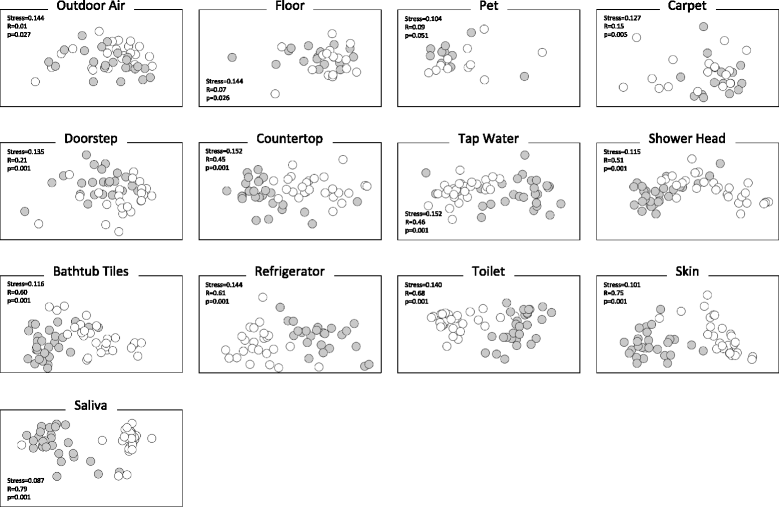
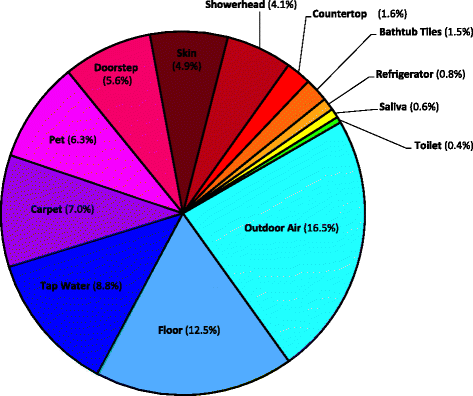
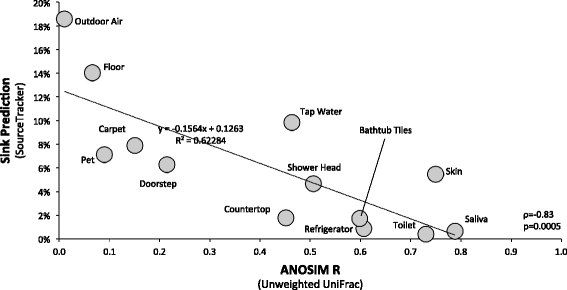
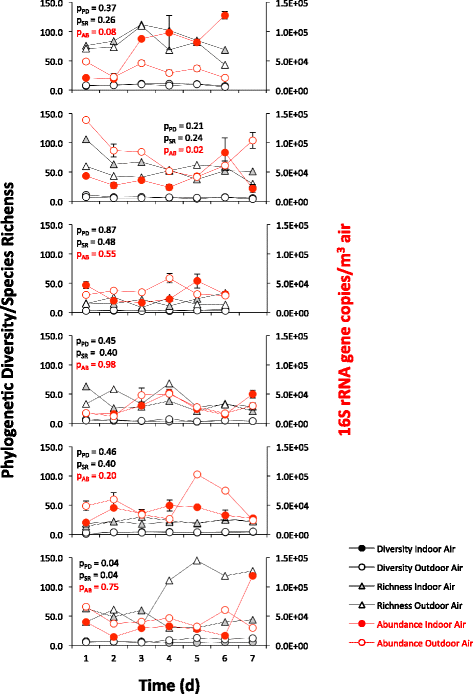
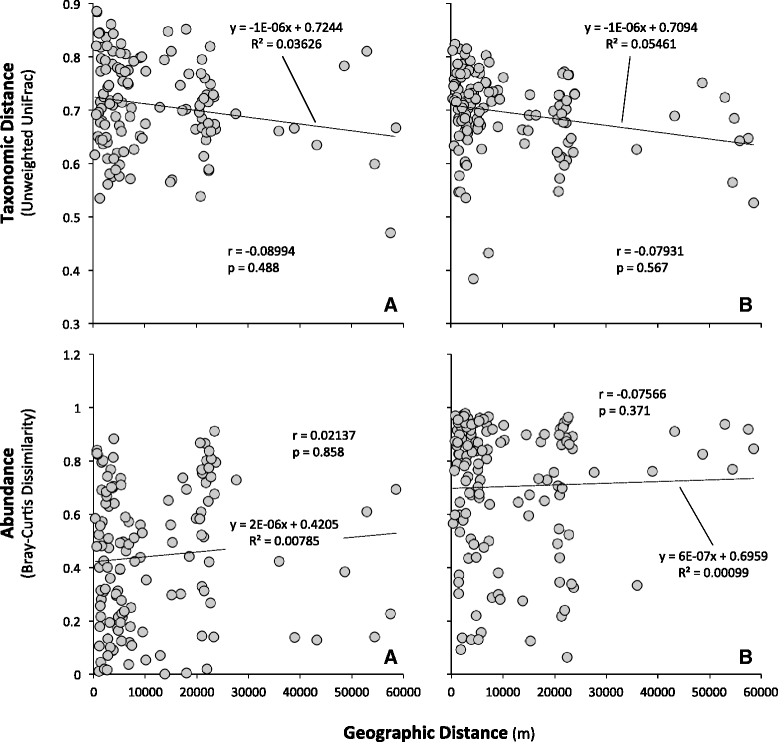
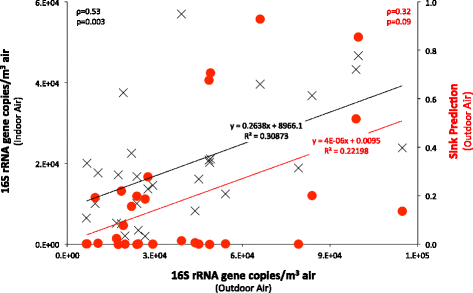
Results: Indoor air harbored a diverse bacterial community dominated by Diaphorobacter sp., Propionibacterium sp., Sphingomonas sp., and Alicyclobacillus sp. Source-sink analysis suggested that outdoor air was the primary source of indoor air microbes in most homes. Bacterial phylogenetic diversity and relative abundance in indoor air did not differ statistically from that in outdoor air. Moreover, the abundance of bacteria in outdoor air was positively correlated with that in indoor air, as would be expected if outdoor air was the main contributor to the bacterial community in indoor bioaerosols. The number of residents, presence of pets, and local tap water also influenced the diversity and size of indoor air microbes. The bacterial load in air increased with the number of residents, activity, and frequency of natural ventilation, and the proportion of bacteria putatively derived from skin increased with the number of residents. Vacuum cleaning increased the signature of pet- and floor-derived bacteria in indoor air, while the frequency of natural ventilation decreased the relative abundance of tap water-derived microorganisms in air.
Conclusions: Indoor air in residences harbors a diverse bacterial community originating from both outdoor and indoor sources and is strongly influenced by household characteristics.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

