Reproducibility of Differential Proteomic Technologies in CPTAC Fractionated Xenografts
- PMID: 26653538
- PMCID: PMC4779376
- DOI: 10.1021/acs.jproteome.5b00859
Reproducibility of Differential Proteomic Technologies in CPTAC Fractionated Xenografts
Abstract
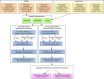
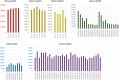
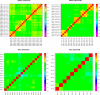
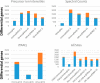
The NCI Clinical Proteomic Tumor Analysis Consortium (CPTAC) employed a pair of reference xenograft proteomes for initial platform validation and ongoing quality control of its data collection for The Cancer Genome Atlas (TCGA) tumors. These two xenografts, representing basal and luminal-B human breast cancer, were fractionated and analyzed on six mass spectrometers in a total of 46 replicates divided between iTRAQ and label-free technologies, spanning a total of 1095 LC-MS/MS experiments. These data represent a unique opportunity to evaluate the stability of proteomic differentiation by mass spectrometry over many months of time for individual instruments or across instruments running dissimilar workflows. We evaluated iTRAQ reporter ions, label-free spectral counts, and label-free extracted ion chromatograms as strategies for data interpretation (source code is available from http://homepages.uc.edu/~wang2x7/Research.htm ). From these assessments, we found that differential genes from a single replicate were confirmed by other replicates on the same instrument from 61 to 93% of the time. When comparing across different instruments and quantitative technologies, using multiple replicates, differential genes were reproduced by other data sets from 67 to 99% of the time. Projecting gene differences to biological pathways and networks increased the degree of similarity. These overlaps send an encouraging message about the maturity of technologies for proteomic differentiation.
Keywords: CPTAC; Differential proteomics; iTRAQ; label-free; quality control; technology assessment; xenografts.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Slebos R. J. C.; Brock J. W. C.; Winters N. F.; Stuart S. R.; Martinez M. A.; Li M.; Chambers M. C.; Zimmerman L. J.; Ham A. J.; Tabb D. L.; et al. Evaluation of strong cation exchange versus isoelectric focusing of peptides for multidimensional liquid chromatography-tandem mass spectrometry. J. Proteome Res. 2008, 7 (12), 5286–5294. 10.1021/pr8004666. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U24-CA-160034/CA/NCI NIH HHS/United States
- U24 CA160034/CA/NCI NIH HHS/United States
- U24-CA-159988/CA/NCI NIH HHS/United States
- U24 CA159988/CA/NCI NIH HHS/United States
- U24-CA-160019/CA/NCI NIH HHS/United States
- P30 CA016086/CA/NCI NIH HHS/United States
- UL1 RR024992/RR/NCRR NIH HHS/United States
- HHSN261201100106C/CA/NCI NIH HHS/United States
- U24 CA160036/CA/NCI NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- U24-CA-160036/CA/NCI NIH HHS/United States
- U24 CA160019/CA/NCI NIH HHS/United States
- UL1 TR000448/TR/NCATS NIH HHS/United States
- U24 CA160035/CA/NCI NIH HHS/United States
- HHSN261201100106C/PHS HHS/United States
- U24-CA-160035/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

