Genome-wide identification and expression profiling of serine proteases and homologs in the diamondback moth, Plutella xylostella (L.)
- PMID: 26653876
- PMCID: PMC4676143
- DOI: 10.1186/s12864-015-2243-4
Genome-wide identification and expression profiling of serine proteases and homologs in the diamondback moth, Plutella xylostella (L.)
Abstract
Background: Serine proteases (SPs) are crucial proteolytic enzymes responsible for digestion and other processes including signal transduction and immune responses in insects. Serine protease homologs (SPHs) lack catalytic activity but are involved in innate immunity. This study presents a genome-wide investigation of SPs and SPHs in the diamondback moth, Plutella xylostella (L.), a globally-distributed destructive pest of cruciferous crops.
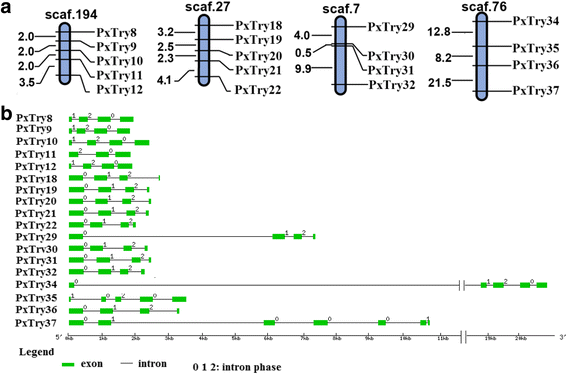
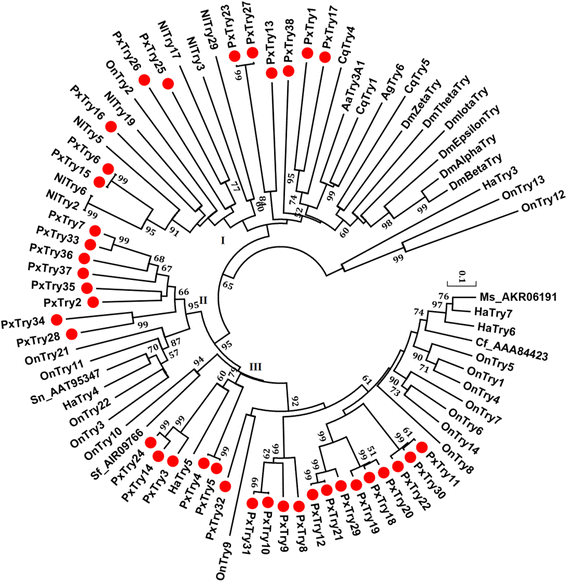
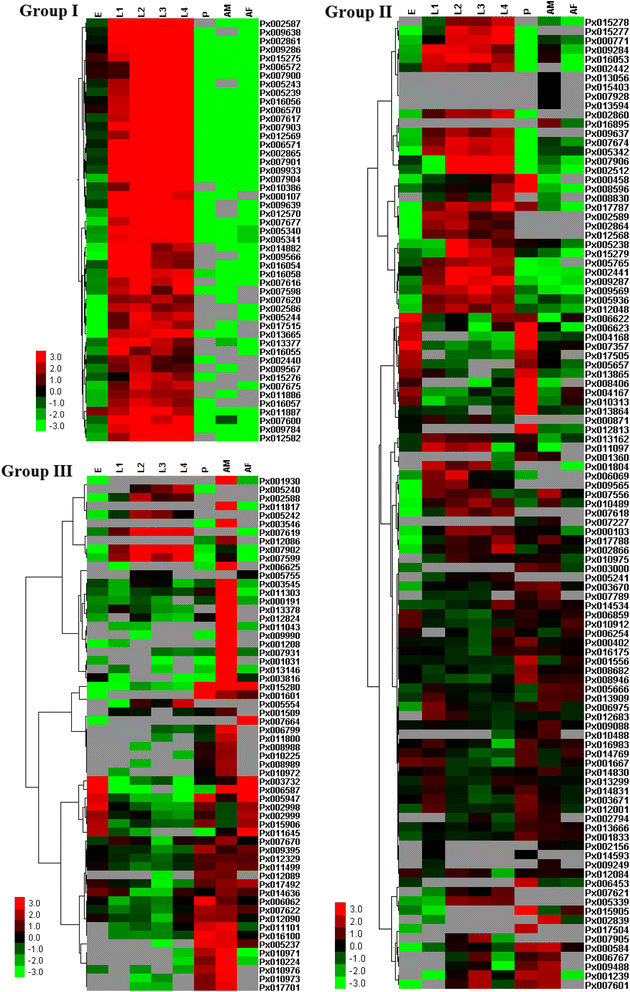
Results: A total of 120 putative SPs and 101 putative SPHs were identified in the P. xylostella genome by bioinformatics analysis. Based on the features of trypsin, 38 SPs were putatively designated as trypsin genes. The distribution, transcription orientation, exon-intron structure and sequence alignments suggested that the majority of trypsin genes evolved from tandem duplications. Among the 221 SP/SPH genes, ten SP and three SPH genes with one or more clip domains were predicted and designated as PxCLIPs. Phylogenetic analysis of CLIPs in P. xylostella, two other Lepidoptera species (Bombyx mori and Manduca sexta), and two more distantly related insects (Drosophila melanogaster and Apis mellifera) showed that seven of the 13 PxCLIPs were clustered with homologs of the Lepidoptera rather than other species. Expression profiling of the P. xylostella SP and SPH genes in different developmental stages and tissues showed diverse expression patterns, suggesting high functional diversity with roles in digestion and development.
Conclusions: This is the first genome-wide investigation on the SP and SPH genes in P. xylostella. The characterized features and profiled expression patterns of the P. xylostella SPs and SPHs suggest their involvement in digestion, development and immunity of this species. Our findings provide a foundation for further research on the functions of this gene family in P. xylostella, and a better understanding of its capacity to rapidly adapt to a wide range of environmental variables including host plants and insecticides.
Figures



Similar articles
-
Characterization and expression profiling of serine protease inhibitors in the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae).BMC Genomics. 2017 Feb 14;18(1):162. doi: 10.1186/s12864-017-3583-z. BMC Genomics. 2017. PMID: 28196471 Free PMC article.
-
Sequence conservation, phylogenetic relationships, and expression profiles of nondigestive serine proteases and serine protease homologs in Manduca sexta.Insect Biochem Mol Biol. 2015 Jul;62:51-63. doi: 10.1016/j.ibmb.2014.10.006. Epub 2014 Dec 19. Insect Biochem Mol Biol. 2015. PMID: 25530503 Free PMC article.
-
Serine protease-related proteins in the malaria mosquito, Anopheles gambiae.Insect Biochem Mol Biol. 2017 Sep;88:48-62. doi: 10.1016/j.ibmb.2017.07.008. Epub 2017 Aug 2. Insect Biochem Mol Biol. 2017. PMID: 28780069 Free PMC article.
-
Drosophila melanogaster clip-domain serine proteases: Structure, function and regulation.Biochimie. 2016 Mar;122:255-69. doi: 10.1016/j.biochi.2015.10.007. Epub 2015 Oct 8. Biochimie. 2016. PMID: 26453810 Review.
-
Serine pseudoproteases in physiology and disease.FEBS J. 2023 May;290(9):2263-2278. doi: 10.1111/febs.16355. Epub 2022 Jan 25. FEBS J. 2023. PMID: 35032346 Review.
Cited by
-
The Draft Genome of Yellow Stem Borer, an Agriculturally Important Pest, Provides Molecular Insights into Its Biology, Development and Specificity Towards Rice for Infestation.Insects. 2021 Jun 19;12(6):563. doi: 10.3390/insects12060563. Insects. 2021. PMID: 34205299 Free PMC article.
-
Hemolymph proteins of Anopheles gambiae larvae infected by Escherichia coli.Dev Comp Immunol. 2017 Sep;74:110-124. doi: 10.1016/j.dci.2017.04.009. Epub 2017 Apr 19. Dev Comp Immunol. 2017. PMID: 28431895 Free PMC article.
-
Identification and gene expression analysis of serine proteases and their homologs in the Asian corn borer Ostrinia furnacalis.Sci Rep. 2023 Mar 23;13(1):4766. doi: 10.1038/s41598-023-31830-2. Sci Rep. 2023. PMID: 36959303 Free PMC article.
-
Genome-wide Identification and Expression Analysis of Amino Acid Transporters in the Whitefly, Bemisia tabaci (Gennadius).Int J Biol Sci. 2017 May 16;13(6):735-747. doi: 10.7150/ijbs.18153. eCollection 2017. Int J Biol Sci. 2017. PMID: 28655999 Free PMC article.
-
RNA Sequencing Reveals the Potential Adaptation Mechanism to Different Hosts of Grapholita molesta.Insects. 2022 Sep 30;13(10):893. doi: 10.3390/insects13100893. Insects. 2022. PMID: 36292841 Free PMC article.
References
-
- Choo YM, Lee KS, Yoon HJ, Lee SB, Kim JH, Sohn HD, et al. A serine protease from the midgut of the bumblebee, Bombus ignites (Hymenoptera: Apidae): cDNA cloning, gene structure, expression and enzyme activity. Eur J Entomol. 2007;104(1):1–7. doi: 10.14411/eje.2007.001. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

