Novel drug targets for personalized precision medicine in relapsed/refractory diffuse large B-cell lymphoma: a comprehensive review
- PMID: 26654227
- PMCID: PMC4676894
- DOI: 10.1186/s12943-015-0474-2
Novel drug targets for personalized precision medicine in relapsed/refractory diffuse large B-cell lymphoma: a comprehensive review
Abstract
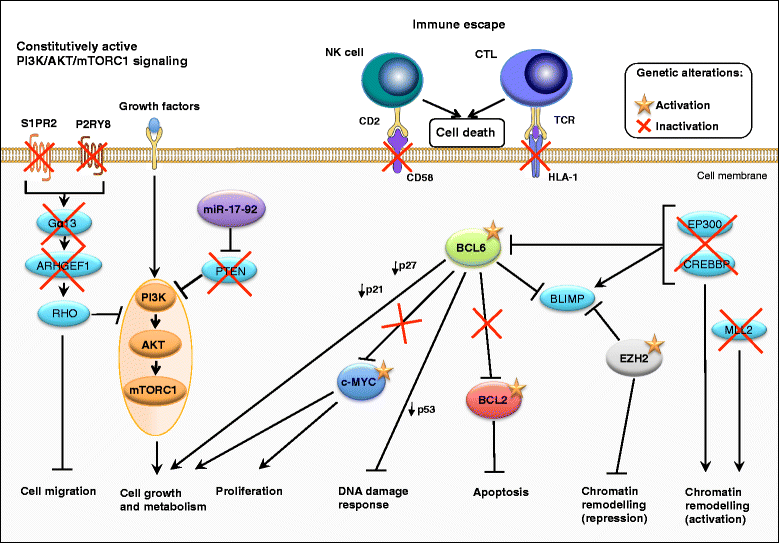
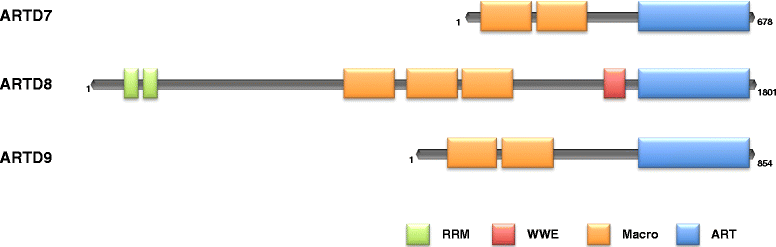
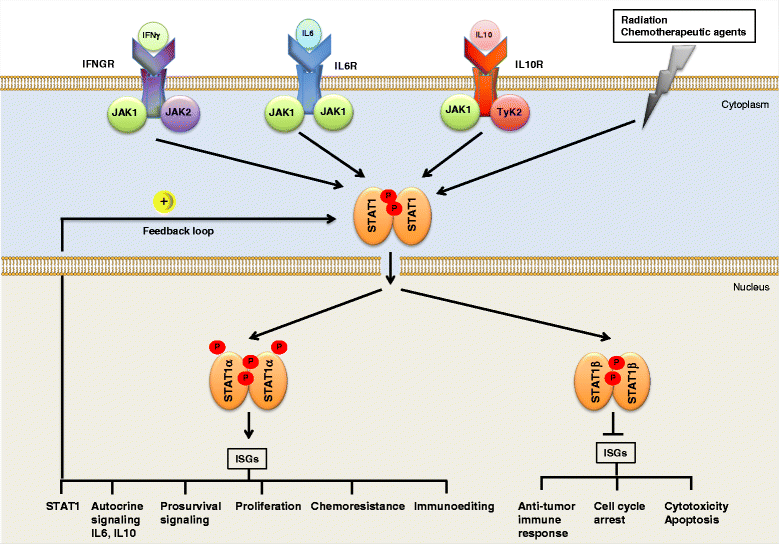
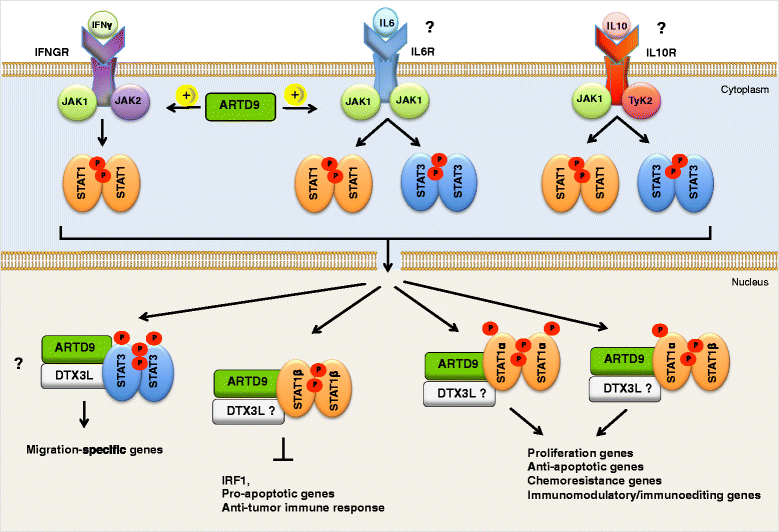
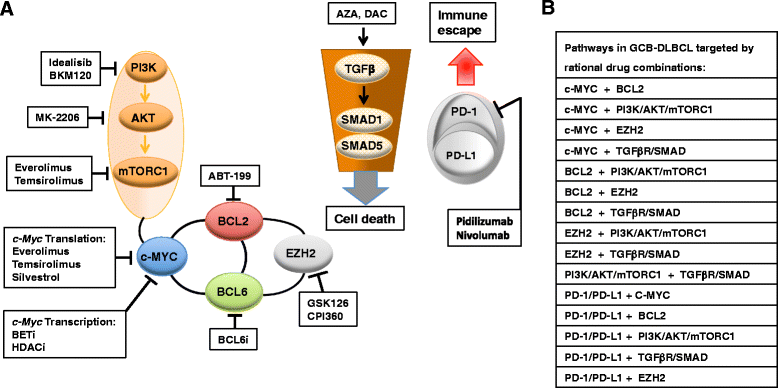
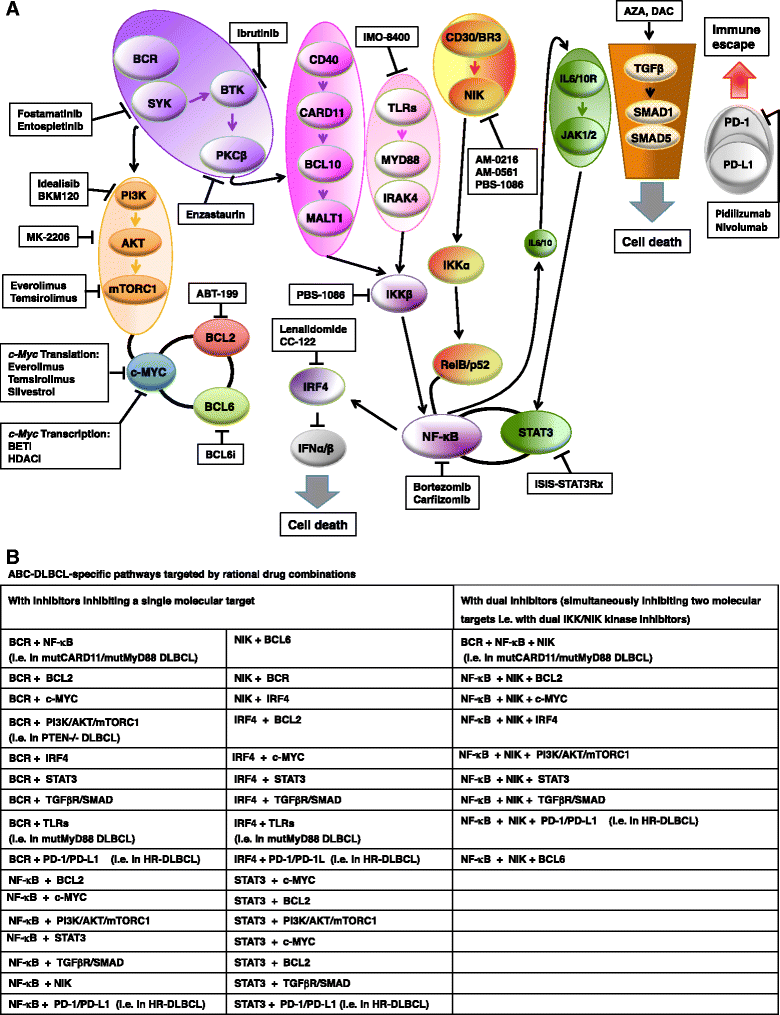
Diffuse large B-cell lymphoma (DLBCL) is a clinically heterogeneous lymphoid malignancy and the most common subtype of non-Hodgkin's lymphoma in adults, with one of the highest mortality rates in most developed areas of the world. More than half of DLBLC patients can be cured with standard R-CHOP regimens, however approximately 30 to 40 % of patients will develop relapsed/refractory disease that remains a major cause of morbidity and mortality due to the limited therapeutic options.Recent advances in gene expression profiling have led to the identification of at least three distinct molecular subtypes of DLBCL: a germinal center B cell-like subtype, an activated B cell-like subtype, and a primary mediastinal B-cell lymphoma subtype. Moreover, recent findings have not only increased our understanding of the molecular basis of chemotherapy resistance but have also helped identify molecular subsets of DLBCL and rational targets for drug interventions that may allow for subtype/subset-specific molecularly targeted precision medicine and personalized combinations to both prevent and treat relapsed/refractory DLBCL. Novel agents such as lenalidomide, ibrutinib, bortezomib, CC-122, epratuzumab or pidilizumab used as single-agent or in combination with (rituximab-based) chemotherapy have already demonstrated promising activity in patients with relapsed/refractory DLBCL. Several novel potential drug targets have been recently identified such as the BET bromodomain protein (BRD)-4, phosphoribosyl-pyrophosphate synthetase (PRPS)-2, macrodomain-containing mono-ADP-ribosyltransferase (ARTD)-9 (also known as PARP9), deltex-3-like E3 ubiquitin ligase (DTX3L) (also known as BBAP), NF-kappaB inducing kinase (NIK) and transforming growth factor beta receptor (TGFβR).This review highlights the new insights into the molecular basis of relapsed/refractory DLBCL and summarizes the most promising drug targets and experimental treatments for relapsed/refractory DLBCL, including the use of novel agents such as lenalidomide, ibrutinib, bortezomib, pidilizumab, epratuzumab, brentuximab-vedotin or CAR T cells, dual inhibitors, as well as mechanism-based combinatorial experimental therapies. We also provide a comprehensive and updated list of current drugs, drug targets and preclinical and clinical experimental studies in DLBCL. A special focus is given on STAT1, ARTD9, DTX3L and ARTD8 (also known as PARP14) as novel potential drug targets in distinct molecular subsets of DLBCL.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

