High-throughput single-molecule screen for small-molecule perturbation of splicing and transcription kinetics
- PMID: 26655523
- PMCID: PMC4766052
- DOI: 10.1016/j.ymeth.2015.11.025
High-throughput single-molecule screen for small-molecule perturbation of splicing and transcription kinetics
Abstract
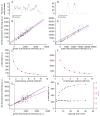
In eukaryotes, mRNA synthesis is catalyzed by RNA polymerase II and involves several distinct steps, including transcript initiation, elongation, cleavage, and transcript release. Splicing of RNA can occur during (co-transcriptional) or after (post-transcriptional) RNA synthesis. Thus, RNA synthesis and processing occurs through the concerted activity of dozens of enzymes, each of which is potentially susceptible to perturbation by small molecules. However, there are few, if any, high-throughput screening strategies for identifying drugs which perturb a specific step in RNA synthesis and processing. Here we have developed a high-throughput fluorescence microscopy approach in single cells to screen for inhibitors of specific enzymatic steps in RNA synthesis and processing. By utilizing the high affinity interaction between bacteriophage capsid proteins (MS2, PP7) and RNA stem loops, we are able to fluorescently label the intron and exon of a β-globin reporter gene in human cells. This approach allows one to measure the kinetics of transcription, splicing and release in both fixed and living cells using a tractable, genetically encoded assay in a stable cell line. We tested this reagent in a targeted screen of molecules that target chromatin readers and writers and identified three compounds that slow transcription elongation without changing transcription initiation.
Keywords: Fluctuation; Fluorescence; RNA; Single-molecule; Small-molecule; Splicing; Transcription.
Published by Elsevier Inc.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

