Selective Recognition of H3.1K36 Dimethylation/H4K16 Acetylation Facilitates the Regulation of All-trans-retinoic Acid (ATRA)-responsive Genes by Putative Chromatin Reader ZMYND8
- PMID: 26655721
- PMCID: PMC4742736
- DOI: 10.1074/jbc.M115.679985
Selective Recognition of H3.1K36 Dimethylation/H4K16 Acetylation Facilitates the Regulation of All-trans-retinoic Acid (ATRA)-responsive Genes by Putative Chromatin Reader ZMYND8
Abstract
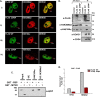
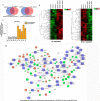
ZMYND8 (zinc finger MYND (Myeloid, Nervy and DEAF-1)-type containing 8), a newly identified component of the transcriptional coregulator network, was found to interact with the Nucleosome Remodeling and Deacetylase (NuRD) complex. Previous reports have shown that ZMYND8 is instrumental in recruiting the NuRD complex to damaged chromatin for repressing transcription and promoting double strand break repair by homologous recombination. However, the mode of transcription regulation by ZMYND8 has remained elusive. Here, we report that through its specific key residues present in its conserved chromatin-binding modules, ZMYND8 interacts with the selective epigenetic marks H3.1K36Me2/H4K16Ac. Furthermore, ZMYND8 shows a clear preference for canonical histone H3.1 over variant H3.3. Interestingly, ZMYND8 was found to be recruited to several developmental genes, including the all-trans-retinoic acid (ATRA)-responsive ones, through its modified histone-binding ability. Being itself inducible by ATRA, this zinc finger transcription factor is involved in modulating other ATRA-inducible genes. We found that ZMYND8 interacts with transcription initiation-competent RNA polymerase II phosphorylated at Ser-5 in a DNA template-dependent manner and can alter the global gene transcription. Overall, our study identifies that ZMYND8 has CHD4-independent functions in regulating gene expression through its modified histone-binding ability.
Keywords: chromatin; chromatin modification; chromatin remodeling; gene expression; gene regulation; gene silencing; gene transcription.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






References
-
- Zhang Y., LeRoy G., Seelig H. P., Lane W. S., and Reinberg D. (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95, 279–289 - PubMed
-
- Tong J. K., Hassig C. A., Schnitzler G. R., Kingston R. E., and Schreiber S. L. (1998) Chromatin deacetylation by an ATP-dependent nucleosome remodeling complex. Nature 395, 917–921 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

