Identification of MEDIATOR16 as the Arabidopsis COBRA suppressor MONGOOSE1
- PMID: 26655738
- PMCID: PMC4703005
- DOI: 10.1073/pnas.1521675112
Identification of MEDIATOR16 as the Arabidopsis COBRA suppressor MONGOOSE1
Abstract
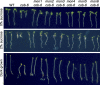
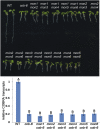
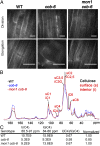
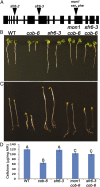
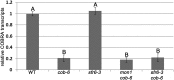
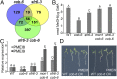
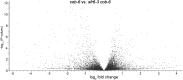
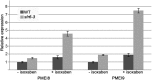
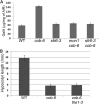
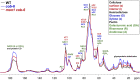
We performed a screen for genetic suppressors of cobra, an Arabidopsis mutant with defects in cellulose formation and an increased ratio of unesterified/esterified pectin. We identified a suppressor named mongoose1 (mon1) that suppressed the growth defects of cobra, partially restored cellulose levels, and restored the esterification ratio of pectin to wild-type levels. mon1 was mapped to the MEDIATOR16 (MED16) locus, a tail mediator subunit, also known as SENSITIVE TO FREEZING6 (SFR6). When separated from the cobra mutation, mutations in MED16 caused resistance to cellulose biosynthesis inhibitors, consistent with their ability to suppress the cobra cellulose deficiency. Transcriptome analysis revealed that a number of cell wall genes are misregulated in med16 mutants. Two of these genes encode pectin methylesterase inhibitors, which, when ectopically expressed, partially suppressed the cobra phenotype. This suggests that cellulose biosynthesis can be affected by the esterification levels of pectin, possibly through modifying cell wall integrity or the interaction of pectin and cellulose.
Keywords: cell wall; cellulose; freezing tolerance; pectin; transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Somerville C, et al. Toward a systems approach to understanding plant cell walls. Science. 2004;306(5705):2206–2211. - PubMed
-
- Wallace IS, Somerville CR. A blueprint for cellulose biosynthesis, deposition, and regulation in plants. In: Fukuda H, editor. Plant Cell Wall Patterning and Cell Shape. Wiley, Hoboken, NJ; 2015. pp. 65–97.
-
- McFarlane HE, Döring A, Persson S. The cell biology of cellulose synthesis. Annu Rev Plant Biol. 2014;65:69–94. - PubMed
-
- Kumar M, Turner S. Plant cellulose synthesis: CESA proteins crossing kingdoms. Phytochemistry. 2015;112:91–99. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

