BmREEPa Is a Novel Gene that Facilitates BmNPV Entry into Silkworm Cells
- PMID: 26656276
- PMCID: PMC4681539
- DOI: 10.1371/journal.pone.0144575
BmREEPa Is a Novel Gene that Facilitates BmNPV Entry into Silkworm Cells
Abstract
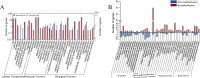
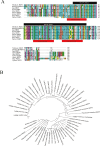
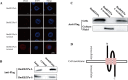
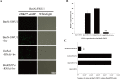
We previously established two silkworm cell lines, BmN-SWU1 and BmN-SWU2, from Bombyx mori ovaries. BmN-SWU1 cells are susceptible while BmN-SWU2 cells are highly resistant to BmNPV infection. Interestingly, we found that the entry of BmNPV into BmN-SWU2 cells was largely inhibited. To explore the mechanism of this inhibition, in this study we used isobaric tags for relative and absolute quantitation (iTRAQ)-based quantitative protein expression profiling and identified 629 differentially expressed proteins between the two cell lines. Among them, we identified a new membrane protein termed BmREEPa. The gene encoding BmREEPa transcribes two splice variants; a 573 bp long BmREEPa-L encoding a protein with 190 amino acids and a 501 bp long BmREEPa-S encoding a protein with 166 amino acids. BmREEPa contains a conserved TB2/DP, HVA22 domain and three transmembrane domains. It is localized in the plasma membrane with a cytoplasmic C-terminus and an extracellular N-terminus. We found that limiting the expression of BmREEPa in BmN-SWU1 cells inhibited BmNPV entry, whereas over-expression of BmREEPa in BmN-SWU2 cells promoted BmNPV entry. Our results also indicated that BmREEPa can interact with GP64, which is the key envelope fusion protein for BmNPV entry. Taken together, the findings of our study revealed that BmREEPa is required for BmNPV to gain entry into silkworm cells, and may provide insights for the identification of BmNPV receptors.
Conflict of interest statement
Figures




Similar articles
-
Differential susceptibilities to BmNPV infection of two cell lines derived from the same silkworm ovarian tissues.PLoS One. 2014 Sep 15;9(9):e105986. doi: 10.1371/journal.pone.0105986. eCollection 2014. PLoS One. 2014. PMID: 25221982 Free PMC article.
-
Proteomic analysis of BmN cell lipid rafts reveals roles in Bombyx mori nucleopolyhedrovirus infection.Mol Genet Genomics. 2017 Apr;292(2):465-474. doi: 10.1007/s00438-016-1284-y. Epub 2017 Feb 3. Mol Genet Genomics. 2017. PMID: 28160078
-
Bombyx mori protein BmREEPa and BmPtchd could form a complex with BmNPV envelope protein GP64.Biochem Biophys Res Commun. 2017 Sep 2;490(4):1254-1259. doi: 10.1016/j.bbrc.2017.07.004. Epub 2017 Jul 3. Biochem Biophys Res Commun. 2017. PMID: 28684317
-
Transgenic RNAi of BmREEPa in silkworms can enhance the resistance of silkworm to Bombyxmori Nucleopolyhedrovirus.Biochem Biophys Res Commun. 2017 Feb 5;483(2):855-859. doi: 10.1016/j.bbrc.2017.01.017. Epub 2017 Jan 6. Biochem Biophys Res Commun. 2017. PMID: 28069383
-
The progress and future of enhancing antiviral capacity by transgenic technology in the silkworm Bombyx mori.Insect Biochem Mol Biol. 2014 May;48:1-7. doi: 10.1016/j.ibmb.2014.02.003. Epub 2014 Feb 20. Insect Biochem Mol Biol. 2014. PMID: 24561307 Review.
Cited by
-
Baculovirus Utilizes Cholesterol Transporter NIEMANN-Pick C1 for Host Cell Entry.Front Microbiol. 2019 Dec 5;10:2825. doi: 10.3389/fmicb.2019.02825. eCollection 2019. Front Microbiol. 2019. PMID: 31866985 Free PMC article.
-
The NPC Families Mediate BmNPV Entry.Microbiol Spectr. 2022 Aug 31;10(4):e0091722. doi: 10.1128/spectrum.00917-22. Epub 2022 Jul 5. Microbiol Spectr. 2022. PMID: 35867410 Free PMC article.
-
Bombyx mori C-Type Lectin (BmIML-2) Inhibits the Proliferation of B. mori Nucleopolyhedrovirus (BmNPV) through Involvement in Apoptosis.Int J Mol Sci. 2022 Jul 28;23(15):8369. doi: 10.3390/ijms23158369. Int J Mol Sci. 2022. PMID: 35955502 Free PMC article.
-
Cross-talking between baculoviruses and host insects towards a successful infection.Philos Trans R Soc Lond B Biol Sci. 2019 Mar 4;374(1767):20180324. doi: 10.1098/rstb.2018.0324. Philos Trans R Soc Lond B Biol Sci. 2019. PMID: 30967030 Free PMC article.
-
Bombyx mori β-1,3-Glucan Recognition Protein 4 (BmβGRP4) Could Inhibit the Proliferation of B. mori Nucleopolyhedrovirus through Promoting Apoptosis.Insects. 2021 Aug 18;12(8):743. doi: 10.3390/insects12080743. Insects. 2021. PMID: 34442307 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

