Natural C-independent expression of restriction endonuclease in a C protein-associated restriction-modification system
- PMID: 26656489
- PMCID: PMC4824078
- DOI: 10.1093/nar/gkv1331
Natural C-independent expression of restriction endonuclease in a C protein-associated restriction-modification system
Abstract
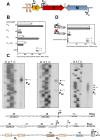
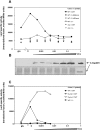
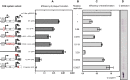
Restriction-modification (R-M) systems are highly prevalent among bacteria and archaea, and appear to play crucial roles in modulating horizontal gene transfer and protection against phage. There is much to learn about these diverse enzymes systems, especially their regulation. Type II R-M systems specify two independent enzymes: a restriction endonuclease (REase) and protective DNA methyltransferase (MTase). Their activities need to be finely balanced in vivo Some R-M systems rely on specialized transcription factors called C (controller) proteins. These proteins play a vital role in the temporal regulation of R-M gene expression, and function to indirectly modulate the horizontal transfer of their genes across the species. We report novel regulation of a C-responsive R-M system that involves a C protein of a poorly-studied structural class - C.Csp231I. Here, the C and REase genes share a bicistronic transcript, and some of the transcriptional auto-control features seen in other C-regulated R-M systems are conserved. However, separate tandem promoters drive most transcription of the REase gene, a distinctive property not seen in other tested C-linked R-M systems. Further, C protein only partially controls REase expression, yet plays a role in system stability and propagation. Consequently, high REase activity was observed after deletion of the entire C gene, and cells bearing the ΔC R-M system were outcompeted in mixed culture assays by those with the WT R-M system. Overall, our data reveal unexpected regulatory variation among R-M systems.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Transcriptome analyses of cells carrying the Type II Csp231I restriction-modification system reveal cross-talk between two unrelated transcription factors: C protein and the Rac prophage repressor.Nucleic Acids Res. 2019 Oct 10;47(18):9542-9556. doi: 10.1093/nar/gkz665. Nucleic Acids Res. 2019. PMID: 31372643 Free PMC article.
-
Natural tuning of restriction endonuclease synthesis by cluster of rare arginine codons.Sci Rep. 2019 Apr 9;9(1):5808. doi: 10.1038/s41598-019-42311-w. Sci Rep. 2019. PMID: 30967604 Free PMC article.
-
Naturally-occurring, dually-functional fusions between restriction endonucleases and regulatory proteins.BMC Evol Biol. 2013 Oct 2;13:218. doi: 10.1186/1471-2148-13-218. BMC Evol Biol. 2013. PMID: 24083337 Free PMC article.
-
Organization and function of the mcrBC genes of Escherichia coli K-12.Mol Microbiol. 1992 May;6(9):1079-86. doi: 10.1111/j.1365-2958.1992.tb01546.x. Mol Microbiol. 1992. PMID: 1316984 Review.
-
ATP-dependent restriction enzymes.Prog Nucleic Acid Res Mol Biol. 2000;64:1-63. doi: 10.1016/s0079-6603(00)64001-1. Prog Nucleic Acid Res Mol Biol. 2000. PMID: 10697406 Review.
Cited by
-
Nonlinear regulatory dynamics of bacterial restriction-modification systems modulates horizontal gene transfer susceptibility.Nucleic Acids Res. 2025 Jan 11;53(2):gkae1322. doi: 10.1093/nar/gkae1322. Nucleic Acids Res. 2025. PMID: 39817515 Free PMC article.
-
Lethal perturbation of an Escherichia coli regulatory network is triggered by a restriction-modification system's regulator and can be mitigated by excision of the cryptic prophage Rac.Nucleic Acids Res. 2024 Apr 12;52(6):2942-2960. doi: 10.1093/nar/gkad1234. Nucleic Acids Res. 2024. PMID: 38153127 Free PMC article.
-
Molecular basis for lethal cross-talk between two unrelated bacterial transcription factors - the regulatory protein of a restriction-modification system and the repressor of a defective prophage.Nucleic Acids Res. 2022 Oct 28;50(19):10964-10980. doi: 10.1093/nar/gkac914. Nucleic Acids Res. 2022. PMID: 36271797 Free PMC article.
-
Regulator-dependent temporal dynamics of a restriction-modification system's gene expression upon entering new host cells: single-cell and population studies.Nucleic Acids Res. 2021 Apr 19;49(7):3826-3840. doi: 10.1093/nar/gkab183. Nucleic Acids Res. 2021. PMID: 33744971 Free PMC article.
-
A transcription factor from the cryptic Escherichia coli Rac prophage controls both phage and host operons.Nucleic Acids Res. 2025 Feb 27;53(5):gkaf113. doi: 10.1093/nar/gkaf113. Nucleic Acids Res. 2025. PMID: 40037713 Free PMC article.
References
-
- Rocha E.P., Danchin A., Viari A. Evolutionary role of restriction/modification systems as revealed by comparative genome analysis. Genome Res. 2001;11:946–958. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

