The Microbiome, Systemic Immune Function, and Allotransplantation
- PMID: 26656674
- PMCID: PMC4771220
- DOI: 10.1128/CMR.00063-15
The Microbiome, Systemic Immune Function, and Allotransplantation
Abstract
Diverse effects of the microbiome on solid organ transplantation are beginning to be recognized. In allograft recipients, microbial networks are disrupted by immunosuppression, nosocomial and community-based infectious exposures, antimicrobial therapies, surgery, and immune processes. Shifting microbial patterns, including acute infectious exposures, have dynamic and reciprocal interactions with local and systemic immune systems. Both individual microbial species and microbial networks have central roles in the induction and control of innate and adaptive immune responses, in graft rejection, and in ischemia-reperfusion injury. Understanding the diverse interactions between the microbiome and the immune system of allograft recipients may facilitate clinical management in the future.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
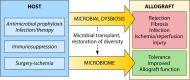
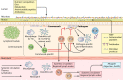


Similar articles
-
Roles of toll-like receptors signaling in organ transplantation.Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2011 Dec;36(12):1125-33. doi: 10.3969/j.issn.1672-7347.2011.12.001. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2011. PMID: 22246357 Review.
-
Innate immunity processes in organ allografting--their contribution to acute and chronic rejection.Ann Transplant. 2005;10(2):5-9. Ann Transplant. 2005. PMID: 16218025 Review.
-
The gut microbiome in solid organ transplantation.Pediatr Transplant. 2020 Nov;24(7):e13866. doi: 10.1111/petr.13866. Epub 2020 Sep 30. Pediatr Transplant. 2020. PMID: 32997434 Review.
-
From the classic concepts to modern practice.Clin Microbiol Infect. 2014 Sep;20 Suppl 7:4-9. doi: 10.1111/1469-0691.12593. Clin Microbiol Infect. 2014. PMID: 24528498 Review.
-
Transplant immunology for non-immunologist.Mt Sinai J Med. 2012 May-Jun;79(3):376-87. doi: 10.1002/msj.21314. Mt Sinai J Med. 2012. PMID: 22678861 Review.
Cited by
-
The Human Virome: Implications for Clinical Practice in Transplantation Medicine.J Clin Microbiol. 2017 Oct;55(10):2884-2893. doi: 10.1128/JCM.00489-17. Epub 2017 Jul 19. J Clin Microbiol. 2017. PMID: 28724557 Free PMC article. Review.
-
Microbiome-based interventions: therapeutic strategies in cancer immunotherapy.Immunooncol Technol. 2020 Nov 28;8:12-20. doi: 10.1016/j.iotech.2020.11.001. eCollection 2020 Dec. Immunooncol Technol. 2020. PMID: 35757563 Free PMC article. Review.
-
Distinct Changes in Gut Microbiota of Patients With Kidney Graft Rejection.Transplant Direct. 2024 Feb 16;10(3):e1582. doi: 10.1097/TXD.0000000000001582. eCollection 2024 Mar. Transplant Direct. 2024. PMID: 38380347 Free PMC article.
-
Case 7-2023: A 70-Year-Old Man with Covid-19, Respiratory Failure, and Rashes.N Engl J Med. 2023 Mar 9;388(10):926-937. doi: 10.1056/NEJMcpc2211369. N Engl J Med. 2023. PMID: 36884326 Free PMC article. No abstract available.
-
2bRAD-M reveals circulating microbiome in chronic antibody-mediated rejection (CAMR) and IgA nephropathy (IgAN) after kidney transplantation.BMC Microbiol. 2025 Jul 31;25(1):468. doi: 10.1186/s12866-025-04103-3. BMC Microbiol. 2025. PMID: 40745634 Free PMC article.
References
-
- Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M. 2009. The NIH Human Microbiome Project. Genome Res 19:2317–2323. doi: 10.1101/gr.096651.109. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

