Scanning Mutagenesis of Human Cytomegalovirus Glycoprotein gH/gL
- PMID: 26656708
- PMCID: PMC4810707
- DOI: 10.1128/JVI.01875-15
Scanning Mutagenesis of Human Cytomegalovirus Glycoprotein gH/gL
Abstract
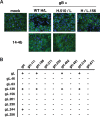
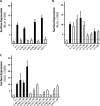
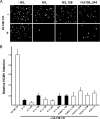
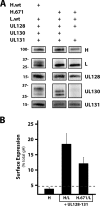
The core, conserved function of the herpesvirus gH/gL is to promote gB-mediated membrane fusion during entry, although the mechanism is poorly understood. The human cytomegalovirus (HCMV) gH/gL can exist as either the gH/gL/gO trimer or the gH/gL/UL128/UL130/UL131 (gH/gL/UL128-131) pentamer. One model suggests that gH/gL/gO provides the core fusion role during entry into all cells within the broad tropism of HCMV, whereas gH/gL/UL128-131 acts at an earlier stage, by a distinct receptor-binding mechanism to enhance infection of select cell types, such as epithelial cells, endothelial cells, and monocytes/macrophages. To further study the distinct functions of these complexes, mutants with individual charged cluster-to-alanine (CCTA) mutations of gH and gL were combined to generate a library of 80 mutant gH/gL heterodimers. The majority of the mutant gH/gL complexes were unable to facilitate gB-mediated membrane fusion in transient-expression cell-cell fusion experiments. In contrast, these mutants supported the formation of gH/gL/UL128-131 complexes that could block HCMV infection in receptor interference experiments. These results suggest that receptor interactions with gH/gL/UL128-131 involve surfaces contained on the UL128-131 proteins but not on gH/gL. gH/gL/UL128-131 receptor interference could be blocked with anti-gH antibodies, suggesting that interference is a cell surface phenomenon and that anti-gH antibodies can block gH/gL/UL128-131 in a manner that is distinct from that for gH/gL/gO.
Importance: Interest in the gH/gL complexes of HCMV (especially gH/gL/UL128-131) as vaccine targets has far outpaced our understanding of the mechanism by which they facilitate entry and contribute to broad cellular tropism. For Epstein-Barr virus (EBV), gH/gL and gH/gL/gp42 are both capable of promoting gB fusion for entry into epithelial cells and B cells, respectively. In contrast, HCMV gH/gL/gO appears to be the sole fusion cofactor that promotes gB fusion activity, whereas gH/gL/UL128-131 expands cell tropism through a distinct yet unknown mechanism. This study suggests that the surfaces of HCMV gH/gL are critical for promoting gB fusion but are dispensable for gH/gL/UL128-131 receptor interaction. This underscores the importance of gH/gL/gO in HCMV entry into all cell types and reaffirms the complex as a candidate target for vaccine development. The two functionally distinct forms of gH/gL present in HCMV make for a useful model with which to study the fundamental mechanisms by which herpesvirus gH/gL regulates gB fusion.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures





References
-
- Britt WJ. 2008. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr Top Microbiol Immunol 325:417–470. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

