Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers
- PMID: 26656844
- PMCID: PMC4857881
- DOI: 10.1038/ng.3470
Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers
Abstract
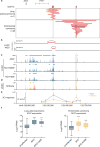
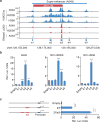
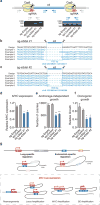
Whole-genome analysis approaches are identifying recurrent cancer-associated somatic alterations in noncoding DNA regions. We combined somatic copy number analysis of 12 tumor types with tissue-specific epigenetic profiling to identify significant regions of focal amplification harboring super-enhancers. Copy number gains of noncoding regions harboring super-enhancers near KLF5, USP12, PARD6B and MYC are associated with overexpression of these cancer-related genes. We show that two distinct focal amplifications of super-enhancers 3' to MYC in lung adenocarcinoma (MYC-LASE) and endometrial carcinoma (MYC-ECSE) are physically associated with the MYC promoter and correlate with MYC overexpression. CRISPR/Cas9-mediated repression or deletion of a constituent enhancer within the MYC-LASE region led to significant reductions in the expression of MYC and its target genes and to the impairment of anchorage-independent and clonogenic growth, consistent with an oncogenic function. Our results suggest that genomic amplification of super-enhancers represents a common mechanism to activate cancer driver genes in multiple cancer types.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

