A new tool called DISSECT for analysing large genomic data sets using a Big Data approach
- PMID: 26657010
- PMCID: PMC4682108
- DOI: 10.1038/ncomms10162
A new tool called DISSECT for analysing large genomic data sets using a Big Data approach
Abstract
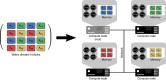
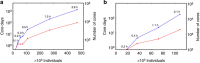
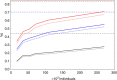
Large-scale genetic and genomic data are increasingly available and the major bottleneck in their analysis is a lack of sufficiently scalable computational tools. To address this problem in the context of complex traits analysis, we present DISSECT. DISSECT is a new and freely available software that is able to exploit the distributed-memory parallel computational architectures of compute clusters, to perform a wide range of genomic and epidemiologic analyses, which currently can only be carried out on reduced sample sizes or under restricted conditions. We demonstrate the usefulness of our new tool by addressing the challenge of predicting phenotypes from genotype data in human populations using mixed-linear model analysis. We analyse simulated traits from 470,000 individuals genotyped for 590,004 SNPs in ∼4 h using the combined computational power of 8,400 processor cores. We find that prediction accuracies in excess of 80% of the theoretical maximum could be achieved with large sample sizes.
Figures

References
-
- Marx V. Biology: the big challenges of big data. Nature 498, 255–260 (2013). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

