Disassembly of the self-assembled, double-ring structure of proteasome α7 homo-tetradecamer by α6
- PMID: 26657688
- PMCID: PMC4677347
- DOI: 10.1038/srep18167
Disassembly of the self-assembled, double-ring structure of proteasome α7 homo-tetradecamer by α6
Abstract
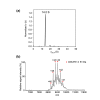
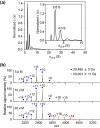
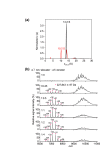
The 20S core particle of the eukaryotic proteasome is composed of two α- and two β-rings, each of which is a hetero-heptamer composed of seven homologous but distinct subunits. Although formation of the eukaryotic proteasome is a highly ordered process assisted by assembly chaperones, α7, an α-ring component, has the unique property of self-assembling into a homo-tetradecamer. We used biophysical methods to characterize the oligomeric states of this proteasome subunit and its interaction with α6, which makes direct contacts with α7 in the proteasome α-ring. We determined a crystal structure of the α7 tetradecamer, which has a double-ring structure. Sedimentation velocity analytical ultracentrifugation and mass spectrometric analysis under non-denaturing conditions revealed that α7 exclusively exists as homo-tetradecamer in solution and that its double-ring structure is disassembled upon the addition of α6, resulting in a 1:7 hetero-octameric α6-α7 complex. Our findings suggest that proteasome formation involves the disassembly of non-native oligomers, which are assembly intermediates.
Figures

Similar articles
-
Two-step process for disassembly mechanism of proteasome α7 homo-tetradecamer by α6 revealed by high-speed atomic force microscopy.Sci Rep. 2017 Nov 13;7(1):15373. doi: 10.1038/s41598-017-15708-8. Sci Rep. 2017. PMID: 29133893 Free PMC article.
-
Mutational and Combinatorial Control of Self-Assembling and Disassembling of Human Proteasome α Subunits.Int J Mol Sci. 2019 May 9;20(9):2308. doi: 10.3390/ijms20092308. Int J Mol Sci. 2019. PMID: 31075988 Free PMC article.
-
Structural Fluctuations of the Human Proteasome α7 Homo-Tetradecamer Double Ring Imply the Proteasomal α-Ring Assembly Mechanism.Int J Mol Sci. 2021 Apr 26;22(9):4519. doi: 10.3390/ijms22094519. Int J Mol Sci. 2021. PMID: 33926037 Free PMC article.
-
Chaperone-driven proteasome assembly.Biochem Soc Trans. 2008 Oct;36(Pt 5):807-12. doi: 10.1042/BST0360807. Biochem Soc Trans. 2008. PMID: 18793141 Review.
-
Comprehensive mass spectrometric analysis of the 20S proteasome complex.Methods Enzymol. 2005;405:187-236. doi: 10.1016/S0076-6879(05)05009-3. Methods Enzymol. 2005. PMID: 16413316 Review.
Cited by
-
Expression, Functional Characterization, and Preliminary Crystallization of the Cochaperone Prefoldin from the Thermophilic Fungus Chaetomium thermophilum.Int J Mol Sci. 2018 Aug 19;19(8):2452. doi: 10.3390/ijms19082452. Int J Mol Sci. 2018. PMID: 30126249 Free PMC article.
-
SDS-induced oligomerization of Lys49-phospholipase A2 from snake venom.Sci Rep. 2019 Feb 20;9(1):2330. doi: 10.1038/s41598-019-38861-8. Sci Rep. 2019. PMID: 30787342 Free PMC article.
-
RecA requires two molecules of Mg2+ ions for its optimal strand exchange activity in vitro.Nucleic Acids Res. 2018 Mar 16;46(5):2548-2559. doi: 10.1093/nar/gky048. Nucleic Acids Res. 2018. PMID: 29390145 Free PMC article.
-
Dynamic structural states of ClpB involved in its disaggregation function.Nat Commun. 2018 Jun 1;9(1):2147. doi: 10.1038/s41467-018-04587-w. Nat Commun. 2018. PMID: 29858573 Free PMC article.
-
A Comprehensive Study of the Interaction between Peptidoglycan Fragments and the Extracellular Domain of Mycobacterium tuberculosis Ser/Thr Kinase PknB.Chembiochem. 2017 Nov 2;18(21):2094-2098. doi: 10.1002/cbic.201700385. Epub 2017 Sep 14. Chembiochem. 2017. PMID: 28851116 Free PMC article.
References
-
- Pickart C. M. Ubiquitin enters the new millennium. Mol Cell 8, 499–504 (2001). - PubMed
-
- Glickman M. H. & Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82, 373–428 (2002). - PubMed
-
- Baumeister W., Walz J., Zühl F. & Seemüller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell 92, 367–80 (1998). - PubMed
-
- Groll M. et al. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature 386, 463–71 (1997). - PubMed
-
- Unno M. et al. The structure of the mammalian 20S proteasome at 2.75 Å resolution. Structure 10, 609–18 (2002). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

