The Epitranscriptome and Innate Immunity
- PMID: 26658668
- PMCID: PMC4675516
- DOI: 10.1371/journal.pgen.1005687
The Epitranscriptome and Innate Immunity
Abstract
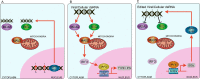
Our knowledge of the variety and abundances of RNA base modifications is rapidly increasing. Modified bases have critical roles in tRNAs, rRNAs, translation, splicing, RNA interference, and other RNA processes, and are now increasingly detected in all types of transcripts. Can new biological principles associated with this diversity of RNA modifications, particularly in mRNAs and long non-coding RNAs, be identified? This review will explore this question by focusing primarily on adenosine to inosine (A-to-I) RNA editing by the adenine deaminase acting on RNA (ADAR) enzymes that have been intensively studied for the past 20 years and have a wide range of effects. Over 100 million adenosine to inosine editing sites have been identified in the human transcriptome, mostly in embedded Alu sequences that form potentially innate immune-stimulating dsRNA hairpins in transcripts. Recent research has demonstrated that inosine in the epitranscriptome and ADAR1 protein establish innate immune tolerance for host dsRNA formed by endogenous sequences. Innate immune sensors that detect viral nucleic acids are among the readers of epitranscriptome RNA modifications, though this does preclude a wide range of other modification effects.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

