Centrosomes are multifunctional regulators of genome stability
- PMID: 26658800
- PMCID: PMC4726469
- DOI: 10.1007/s10577-015-9506-4
Centrosomes are multifunctional regulators of genome stability
Abstract
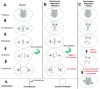
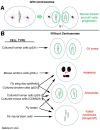
The maintenance of genome stability is critical for proper cell function, and loss of this stability contributes to many human diseases and developmental disorders. Therefore, cells have evolved partially redundant mechanisms to monitor and protect the genome. One subcellular organelle implicated in the maintenance of genome stability is the centrosome, best known as the primary microtubule organizing center of most animal cells. Centrosomes serve many different roles throughout the cell cycle, and many of those roles, including mitotic spindle assembly, nucleation of the interphase microtubule array, DNA damage response, and efficient cell cycle progression, have been proposed to help maintain genome stability. As a result, the centrosome is itself a highly regulated entity. Here, we review evidence concerning the significance of the centrosome in promoting genome integrity. Recent advances permitting acute and persistent centrosome removal suggest we still have much to learn regarding the specific function and actual importance of centrosomes in different contexts, as well as how cells may compensate for centrosome dysfunction to maintain the integrity of the genome. Although many animal cells survive and proliferate in the absence of centrosomes, they do so aberrantly. Based on these and other studies, we conclude that centrosomes serve as critical, multifunctional organelles that promote genome stability.
Keywords: Acentrosomal; Aneuploidy; Asymmetric division; Cell cycle; Centrosome; Centrosome separation; Chromosomal instability; DNA damage; Genome stability; Interphase; Mitosis; PCM; p53.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

