ENIGMA and the individual: Predicting factors that affect the brain in 35 countries worldwide
- PMID: 26658930
- PMCID: PMC4893347
- DOI: 10.1016/j.neuroimage.2015.11.057
ENIGMA and the individual: Predicting factors that affect the brain in 35 countries worldwide
Abstract
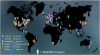
In this review, we discuss recent work by the ENIGMA Consortium (http://enigma.ini.usc.edu) - a global alliance of over 500 scientists spread across 200 institutions in 35 countries collectively analyzing brain imaging, clinical, and genetic data. Initially formed to detect genetic influences on brain measures, ENIGMA has grown to over 30 working groups studying 12 major brain diseases by pooling and comparing brain data. In some of the largest neuroimaging studies to date - of schizophrenia and major depression - ENIGMA has found replicable disease effects on the brain that are consistent worldwide, as well as factors that modulate disease effects. In partnership with other consortia including ADNI, CHARGE, IMAGEN and others1, ENIGMA's genomic screens - now numbering over 30,000 MRI scans - have revealed at least 8 genetic loci that affect brain volumes. Downstream of gene findings, ENIGMA has revealed how these individual variants - and genetic variants in general - may affect both the brain and risk for a range of diseases. The ENIGMA consortium is discovering factors that consistently affect brain structure and function that will serve as future predictors linking individual brain scans and genomic data. It is generating vast pools of normative data on brain measures - from tens of thousands of people - that may help detect deviations from normal development or aging in specific groups of subjects. We discuss challenges and opportunities in applying these predictors to individual subjects and new cohorts, as well as lessons we have learned in ENIGMA's efforts so far.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Adams H, the CHARGE and ENIGMA2 Consortia. Adams Dr. Hieab, Hibar Dr. Derrek, Chouraki Dr. Vincent, Stein Dr. Jason, Nyquist Dr. Paul, Renteria Dr. Miguel, Trompet Dr. Stella, Arias-Vasquez Dr. Alejandro, Seshadri Dr. Sudha, Desrivieres Dr. Sylvane, Beecham Dr. Ashley, Jahanshad Dr. Neda, Wittfeld Dr. Katharina, Van der Lee Dr. Sven, Abramovic Ms. Lucija, Alhusaini Dr. Saud, Amin Dr. Najaf, Andersson Dr. Micael, Arfanakis Dr. Konstantinos, Aribisala Dr. Benjamin, Armstrong Dr. Nicola, Athanasiu Lavinia, Axelsson Dr. Tomas, Beiser Dr. Alexa, Bernard Ms. Manon, Bis Dr. Joshua, Blanken Dr. Laura, Blanton Dr. Susan, Bohlken Mr. Marc, Boks Dr. Marco, Bralten Dr. Janita, Brickman Dr. Adam, Carmichael Dr. Owen, Chakravarty Dr. Mallar, Chauhan Dr. Ganesh, Chen Dr. Qiang, Ching Dr. Christopher, Cuellar-Partida Dr. Gabriel, den Braber Dr. Anouk, Trung Doan Dr. Nhat, Ehrlich Dr. Stefan, Filippi Dr. Irina, Ge Tian, Giddaluru Dr. Sudheer, Goldman Dr. Aaron, Gottesman Dr. Rebecca, Greven Dr. Corina, Grimm Dr. Oliver, Griswold Dr. Michael, Guadalupe Dr. Tulio, Hass Johanna, Haukvik Unn, Hilal Dr. Saima, Hofer Dr. Edith, Hoehn Dr. David, Holmes Dr. Avram, Hoogman Dr. Martine, Janowitz Dr. Deborah, Jia Dr. Tianye, Karbalai Dr. Nazanin, Kasperaviciute Dr. Dalia, Kim Dr. Sungeun, Marieke Klein Miss, Kraemer Mr. Bernd, Lee Dr. Phil, Liao Dr. Jiemin, Liewald Mr. David, Lopez Dr. Lorna, Luciano Dr. Michelle, Macare Ms. Christine, Marquand Dr. Andre, Matarin Dr. Mar, Mather Dr. Karen, Mattheisen Manuel, Mazoyer Dr. Bernard, McKay Dr. David, McWhirter Dr. Rebekah, Milaneschi Dr. Yuri, Muetzel Dr. Ryan, Muñoz Maniega Dr. Susana, Nho Dr. Kwangsik, Nugent Dr. Allison, Loohuis Dr. Loes Olde, Oosterlaan Dr. Jaap, Papmeyer Dr. Martina, Pappa Dr. Irene, Pirpamer Dr. Lukas, Pudas Dr. Sara, Pütz Dr. Benno, Rajan Dr. Kumar, Ramasamy Dr. Adaikalavan, Richards Dr. Jennifer, Risacher Dr. Shannon, Roiz-Santiañez Dr. Roberto, Rommelse Dr. Nanda, Rose Dr. Emma, Natalie Royle Miss, Rundek Dr. Tatjana, Sämann Dr. Philipp, Satizabal Dr. Claudia, Schmaal Dr. Lianne, Schork Mr. Andrew, Shen Dr. Li, Shin Dr. Jean, Shumskaya Dr. Elena, Smith Dr. Albert, Sprooten Dr. Emma, Strike Dr. Lachlan, Teumer Dr. Alexander, Thomson Dr. Russell, Tordesillas-Gutierrez Dr. Diana, Toro Mr. Roberto, Trabzuni Dr. Daniah, Vaidya Dr. Dhananjay, Van der Grond Dr. Jeroen, Van der Meer Dr. Dennis, Van Donkelaar Dr. Marjolein, Van Eijk Dr. Kristel, van Erp Dr. Theo, Van Rooij Dr. Daan, Walton Esther, Tjelta Westlye Dr. Lars, Whelan Dr. Christopher, Windham Dr. Beverly, Winkler Dr. Anderson, Woldehawariat Dr. Girma, Wolf Dr. Christiane, Wolfers Dr. Thomas, Xu Dr. Bing, Yanek Dr. Lisa, Yang Dr. Jingyun, Zijdenbos Dr. Alex, Zwiers Dr. Marcel, Agartz Ms. Ingrid, Aggarwal Dr. Neelum, Almasy Dr. Laura, Ames Dr. David, Amouyel Philippe, Andreassen Prof. Ole, Arepalli Dr. Sampath, Assareh Amelia, Barral Dr. Sandra, Bastin Dr. Mark, Becker Dr. James, Becker Dr. Diane, Bennett Dr. David, Blangero Dr. John, Bokhoven Dr. Hans, Boomsma Dr. Dorret, Brodaty Prof. Henry, Brouwer Dr. Rachel, Brunner Prof. Han, Buckner Dr. Randy, Buitelaar Dr. Jan, Bulayeva Dr. Kazima, Cahn Mrs. Wiepke, Calhoun Dr. Vince, Cannon Dara, Cavalleri Dr. Gianpiero, Chen Dr. Christopher, Cheng Dr. Ching-Yu, Cichon Prof. Sven, Cookson Dr. Mark, Corvin Dr. Aiden, Crespo-Facorro Benedicto, Curran Dr. Joanne, Czisch Michael, Dale Dr. Anders, Davies Dr. Gareth, de Geus Prof. Eco, De Jager Dr. Philip, De Zubicaray Dr. Greig, Delanty Dr. Norman, Depondt Dr. Chantal, DeStefano Dr. Anita, Dillman Dr. Allissa, Djurovic Dr. Srdjan, Donohoe Dr. Gary, Drevets Dr. Wayne, Duggirala Dr. Ravi, Dyer Dr. Thomas, Erk Dr. Susanne, Espeseth Dr. Thomas, Evans Dr. Denis, Fedko Dr. Iryna, Fernández Guillén, Ferrucci Dr. Luigi, Fisher Prof. Simon, Fleischman Dr. Debra, Ford Dr. Ian, Foroud Dr. Tatiana, Fox Dr. Peter, Francks Dr. Clyde, Fukunaga Masaki, Gibbs J, Glahn Dr. David, Gollub Dr. Randy, Göring Dr. Harald, Grabe Dr. Hans, Green Dr. Robert, Gruber Dr. Oliver, Guelfi Mr. Manuel, Hansell Dr. Narelle, Hardy John, Hartman Dr. Catharina, Hashimoto Dr. Ryota, Hegenscheid Dr. Katrin, Heinz Dr. Andreas, Hellard Dr. Stephanie, Hernandez Dr. Dena, Heslenfeld Dr. Dirk, Ho Dr. Beng-Choon, Hoekstra Prof. Pieter, Hoffmann Dr. Wolfgang, Hofman Prof. Albert, Holsboer Dr. Florian, Homuth Dr. Georg, Hosten Dr. Norbert, Hottenga Dr. Jouke, Hulshoff Pol Dr. Hilleke, Ikeda Dr. Masashi, Ikram Dr. M Kamran, Jack Dr. Clifford, Jenkinson Dr. Mark, Johnson Dr. Robert, Jonsson Erik, Jukema Prof. J Wouter, Kahn Dr. Rene, Kanai Ryota, Kloszewska Dr. Iwona, Knopman David, Kochunov Dr. Peter, Kwok John, Launer Dr. Lenore, Lawrie Dr. Stephen, Lemaître Hervé, Liu Dr. Xinmin, Longo Dr. Dan, Longstreth Dr. WT, Jr, Lopez Dr. Oscar, Lovestone Dr. Simon, Martinez Dr. Oliver, Martinot Dr. Jean-Luc, Mattay Venkata, McDonald, Prof Prof. Colm, McIntosh Andrew, McMahon Dr. Francis, McMahon Dr. Katie, mecocci Prof. patrizia, Melle Dr. Ingrid, Meyer-Lindenberg Prof. Andreas, Mohnke Mr. Sebastian, Montgomery Dr. Grant, Morris Dr. Derek, Mosley Dr. Thomas, Mühleisen Dr. Thomas, Müller-Myhsok Dr. Bertram, Nalls Dr. Michael, Nauck Dr. Matthias, Nichols Dr. Thomas, Niessen, Prof Prof. Wiro, Nöthen Markus, Nyberg Prof. Lars, Ohi Dr. Kazutaka, Olvera Dr. Rene, Ophoff Roel, Pandolfo Dr. Massimo, Paus Dr. Tomas, Pausova Dr. Zdenka, Penninx Prof. Brenda, Pike Dr. G Bruce, Potkin Prof. Steven, Psaty Dr. Bruce, Reppermund Dr. Simone, Rietschel Prof. Marcella, Roffman Dr. Joshua, Romanczuk-Seiferth Dr. Nina, Rotter Dr. Jerome, Ryten Dr. Mina, Sacco Dr. Ralph, Sachdev Prof. Perminder, Saykin Dr. Andrew, Schmidt Dr. Reinhold, Schofield Dr. Peter, Sigursson Dr. Sigurdur, Simmons Dr. Andrew, Singleton Dr. Andrew, Sisodiya Prof. Sanjay, Smith Dr. Colin, Smoller Dr. Jordan, Soininen Prof. Hilkka, Srikanth Dr. Velandai, Steen Dr. Vidar, Stott Dr. David, Sussmann Jess, Thalamuthu Dr. Anbupalam, Tiemeier Dr. Henning, Toga Dr. Arthur, Traynor Dr. Bryan, Troncoso Dr. Juan, Turner Jessica, Tzourio Dr. Christophe, Uitterlinden André, Valdés Hernández Dr. Maria, Van der Brug Dr. Marcel, van der Lugt Prof. Aad, van der Wee Dr. Nic, van Duijn Prof. Cornelia, van Haren Dr. Neeltje, van 't Ent Dr. Dennis, Van Tol Dr. Marie-Jose, Vardarajan Dr. Badri, Veltman Dr. Dick, Vernooij Dr. Meike, Völzke Dr. Henry, Walter Henrik, Wardlaw Prof. Joanna, Wassink Dr. Thomas, Weale Mike, Weinberger Dr. Daniel, Weiner Prof. Michael, Wen Dr. Wei, Westman Dr. Eric, White Dr. Tonya, Wong Dr. Tien, Wright Dr. Clinton, Dr. Ronal Common genetic variation underlying human intracranial volume highlights developmental influences and continued relevance during late life. 2015 submitted for publication, October 2015.
-
- Ahmed M, Cannon DM, Scanlon C, Holleran L, Schmidt H, McFarland J, Langan C, McCarthy P, Barker GJ, Hallahan B, McDonald C. Progressive brain atrophy and cortical thinning in schizophrenia after commencing clozapine treatment. Neuropsychopharmacology. 2015. (Apr 1, Epub ahead of print) - DOI - PMC - PubMed
-
- Aristotle (350 BCE), d. Metaphysics α. available online at http://www.isnature.org/Files/Aristotle/.
-
- Ashbrook DG, Williams RW, Lu L, Stein JL, Hibar DP, Nichols TE, Medland SE, Thompson PM, Hager R. Joint genetic analysis of hippocampal size in mouse and human identifies a novel gene linked to neurodegenerative disease. BMC Genomics. 2014;15:850. doi: 10.1186/1471-2164-15-850. (Oct 3) - DOI - PMC - PubMed
-
- Backhouse EV, McHutchison CA, Cvoro V, Shenkin SD, Wardlaw JM. Early life risk factors for stroke and cognitive impairment. Curr. Epidemiol. Rep. 2015. - DOI
Publication types
MeSH terms
Grants and funding
- RF1 AG041915/AG/NIA NIH HHS/United States
- R01 AG018386/AG/NIA NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- R01 MH104284/MH/NIMH NIH HHS/United States
- U54 EB020403/EB/NIBIB NIH HHS/United States
- G0900908/Medical Research Council/United Kingdom
- R00 MH101367/MH/NIMH NIH HHS/United States
- P01 AA019072/AA/NIAAA NIH HHS/United States
- G0701120/Medical Research Council/United Kingdom
- BB/F019394/1/Biotechnology and Biological Sciences Research Council/United Kingdom
- K24 MH094614/MH/NIMH NIH HHS/United States
- K99 MH101367/MH/NIMH NIH HHS/United States
- R01 EB015611/EB/NIBIB NIH HHS/United States
- R01 AG040060/AG/NIA NIH HHS/United States
- MR/M013111/1/Medical Research Council/United Kingdom
- R01 AG018384/AG/NIA NIH HHS/United States
- G1001245/Medical Research Council/United Kingdom
- P30 AG028740/AG/NIA NIH HHS/United States
- 100309/Wellcome Trust/United Kingdom
- R01 DA047119/DA/NIDA NIH HHS/United States
- MR/K026992/1/Medical Research Council/United Kingdom
- R01 AG022381/AG/NIA NIH HHS/United States
- R01 AG050595/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

