Circular RNA enrichment in platelets is a signature of transcriptome degradation
- PMID: 26660425
- PMCID: PMC4797142
- DOI: 10.1182/blood-2015-06-649434
Circular RNA enrichment in platelets is a signature of transcriptome degradation
Abstract
In platelets, splicing and translation occur in the absence of a nucleus. However, the integrity and stability of mRNAs derived from megakaryocyte progenitor cells remain poorly quantified on a transcriptome-wide level. As circular RNAs (circRNAs) are resistant to degradation by exonucleases, their abundance relative to linear RNAs can be used as a surrogate marker for mRNA stability in the absence of transcription. Here we show that circRNAs are enriched in human platelets 17- to 188-fold relative to nucleated tissues and 14- to 26-fold relative to samples digested with RNAse R to selectively remove linear RNA. We compare RNAseq read depths inside and outside circRNAs to provide in silico evidence of transcript circularity, show that exons within circRNAs are enriched on average 12.7 times in platelets relative to nucleated tissues and identify 3162 genes significantly enriched for circRNAs, including some where all RNAseq reads appear to be derived from circular molecules. We also confirm that this is a feature of other anucleate cells through transcriptome sequencing of mature erythrocytes, demonstrate that circRNAs are not enriched in cultured megakaryocytes, and demonstrate that linear RNAs decay more rapidly than circRNAs in platelet preparations. Collectively, these results suggest that circulating platelets have lost >90% of their progenitor mRNAs and that translation in platelets occurs against the backdrop of a highly degraded transcriptome. Finally, we find that transcripts previously classified as products of reverse transcriptase template switching are both enriched in platelets and resistant to decay, countering the recent suggestion that up to 50% of rearranged RNAs are artifacts.
© 2016 by The American Society of Hematology.
Figures
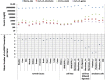

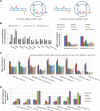
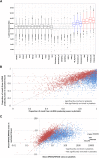
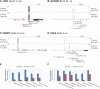
Comment in
-
Platelets enrich their transcriptome circle.Blood. 2016 Mar 3;127(9):1080-1. doi: 10.1182/blood-2015-12-687723. Blood. 2016. PMID: 26941389
References
-
- Deutsch VR, Tomer A. Megakaryocyte development and platelet production. Br J Haematol. 2006;134(5):453–466. - PubMed
-
- Harker LA, Roskos LK, Marzec UM, et al. Effects of megakaryocyte growth and development factor on platelet production, platelet life span, and platelet function in healthy human volunteers. Blood. 2000;95(8):2514–2522. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

