Reprogramming the genetic code: The emerging role of ribosomal frameshifting in regulating cellular gene expression
- PMID: 26661048
- PMCID: PMC4749135
- DOI: 10.1002/bies.201500131
Reprogramming the genetic code: The emerging role of ribosomal frameshifting in regulating cellular gene expression
Abstract
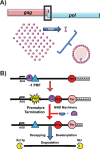
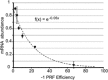
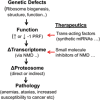
Reading frame maintenance is a critical property of ribosomes. However, a number of genetic elements have been described that can induce ribosomes to shift on mRNAs, the most well understood of which are a class that directs ribosomal slippage by one base in 5' (-1) direction. This is referred to as programmed -1 ribosomal frameshifting (-1 PRF). Recently, a new -1 PRF promoting element was serendipitously discovered in a study examining the effects of stretches of adenosines in the coding sequences of mRNAs. Here, we discuss this finding, recent studies describing how -1 PRF is used to control gene expression in eukaryotes, and how -1 PRF is itself regulated. The implications of dysregulation of -1 PRF on human health are examined, as are possible new areas in which novel -1 PRF promoting elements might be discovered. Also watch the Video Abstract.
Keywords: NMD; SCA26; cancer; frameshifting; miRNA; polyA track; pseudoknot; ribosome; ribosomopathy; telomere; translation.
© 2015 WILEY Periodicals, Inc.
Figures
References
-
- Cochella L, Green R. 2005. Fidelity in protein synthesis. Curr Biol 15: R536–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

